Algorithm and FIJI plugins developed by Hideo Otsuna.
This program is designed for automatic 2D segmentation of zebrafish in bright-field microscopy images. It also automatically segments fluorescent signals within the zebrafish, measuring both the signal pixel area and its brightness.
- Download .zip file from here Zebrafish 96-well segmentation measure FPC release. And unzip the file.
- Launch FIJI and ensure it's updated.
- Copy "Zebra_96well.ijm" to /Fiji.app/plugins/Macros/
- Place "Size_based_Noise_elimination.jar", "Gamma_samewindow_noswing.jar", and "Local_contrast_thresholding.jar" in /Fiji.app/plugins/
- Restart Fiji.
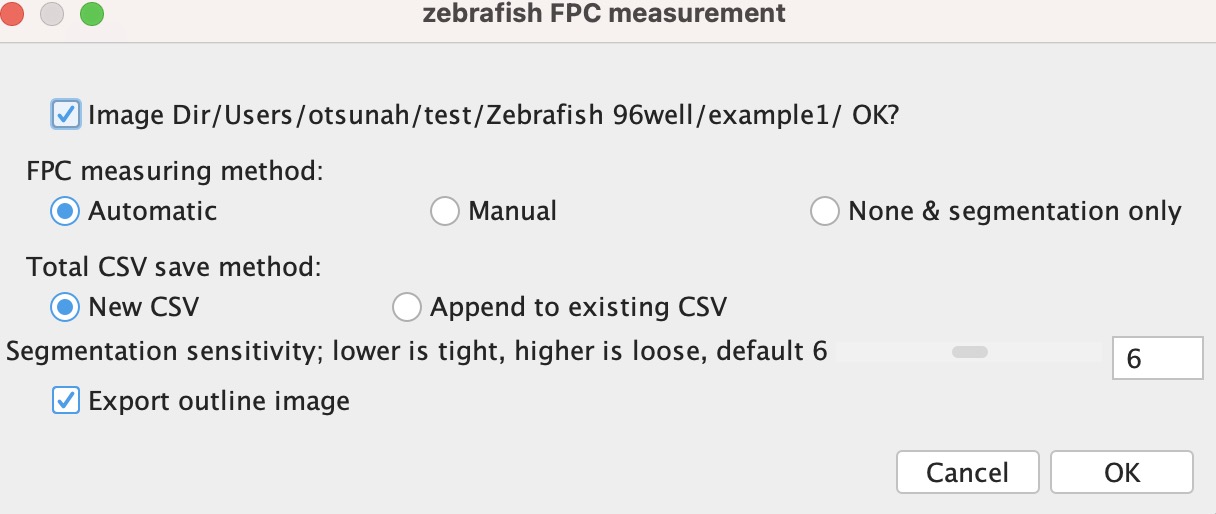
Select menu: Plugins/Macros/Zebra_96well
• Image Dir: When unchecked, the program prompts you to specify the folder containing the 2D zebrafish images.
• FPC measuring method:
- Automatic: This mode will automatically segment the fluorescent signals.
- Manual: This mode requires manual input for segmentation.
- None & segmentation only: Outputs only the segmented zebrafish images.
• Total CSV save method: The program saves a .csv file one directory up from the "Image Dir." This .csv will contain measurement data such as "Sample name, Sum area, Averaged area, Sum brightness, Averaged brightness, Number of FPC" for all images.
- New CSV: Generates a new .csv named after the folder.
- Append to existing CSV: Appends the measurement data to an existing .csv named after the folder.
• Segmentation sensitivity: A lower value will segment the zebrafish with a tighter border. A higher value allows more space around the zebrafish. If the segmentation results in a chopped zebrafish, adjust to a larger value.
• Export outline image: When enabled, this will output an image showing the outline of the segmented zebrafish.