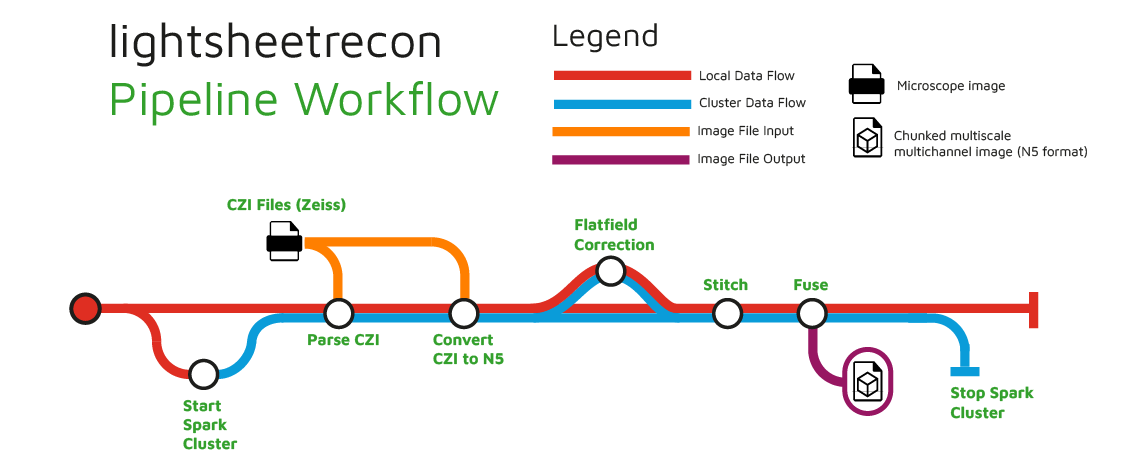
nf-core/lightsheetrecon is a bioimage analysis pipeline that reconstructs large microscopy image volumes. It ingests raw images in CZI format from Zeiss Lightsheet microscopes, computes flatfield correction and tile stitching, and outputs a multi-resolution image pyramid in N5 format. In the future this pipeline will support additional input formats as well as deconvolution and other image processing methods.
- Spin up a Spark cluster if configured
- Read image metadata from MVL metadata file (stitching-spark)
- Convert the CZI images to N5 format (stitching-spark)
- Compute flatfield correction (stitching-spark)
- Compute stitching (stitching-spark)
- Fuse files and export to N5 (stitching-spark)
- Stop Spark cluster if configured
Note If you are new to Nextflow and nf-core, please refer to this page on how to set-up Nextflow. Make sure to test your setup with
-profile testbefore running the workflow on actual data.
First, prepare a samplesheet with your input data that looks as follows:
samplesheet.csv:
id,filename,pattern
LHA3_R3_tiny,LHA3_R3_small.czi,LHA3_R3_small.czi
LHA3_R3_tiny,LHA3_R3_tiny.mvl,
LHA3_R5_tiny,LHA3_R5_small.czi,LHA3_R5_small.czi
LHA3_R5_tiny,LHA3_R5_tiny.mvl,
Each row represents a file in the input data set. The identifier (id) groups files together into acquisition rounds. In the example above, each acquisition is a single CZI file containing all of the tiles and channels, and an MVL file containing the acquisition metadata (e.g. stage coordinates for each tile.)
Now, you can run the pipeline using:
nextflow run nf-core/lightsheetrecon \
-profile <docker/singularity/.../institute> \
--input samplesheet.csv \
--outdir <OUTDIR>:::warning
Please provide pipeline parameters via the CLI or Nextflow -params-file option. Custom config files including those
provided by the -c Nextflow option can be used to provide any configuration except for parameters;
see docs.
:::
For more details and further functionality, please refer to the usage documentation and the parameter documentation.
To see the results of an example test run with a full size dataset refer to the results tab on the nf-core website pipeline page. For more details about the output files and reports, please refer to the output documentation.
The stitching-spark tools used in this pipeline were developed by the Saalfeld Lab at Janelia Research Campus.
The modules for running Spark clusters on Nextflow were originally prototyped by Cristian Goina.
The nf-core/lightsheetrecon pipeline was originally constructed by Konrad Rokicki.
The workflow diagram is based on the SVG source from the cutandrun pipeline.
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don't hesitate to get in touch on the Slack #lightsheetrecon channel (you can join with this invite).
If you use nf-core/lightsheetrecon for your analysis, please cite the EASI-FISH article as follows:
Yuhan Wang, Mark Eddison, Greg Fleishman, Martin Weigert, Shengjin Xu, Fredrick E. Henry, Tim Wang, Andrew L. Lemire, Uwe Schmidt, Hui Yang, Konrad Rokicki, Cristian Goina, Karel Svoboda, Eugene W. Myers, Stephan Saalfeld, Wyatt Korff, Scott M. Sternson, Paul W. Tillberg. Expansion-Assisted Iterative-FISH defines lateral hypothalamus spatio-molecular organization. Cell. 2021 Dec 22;184(26):6361-6377.e24. doi: 10.1016/j.cell.2021.11.024. PubMed PMID: 34875226.
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.