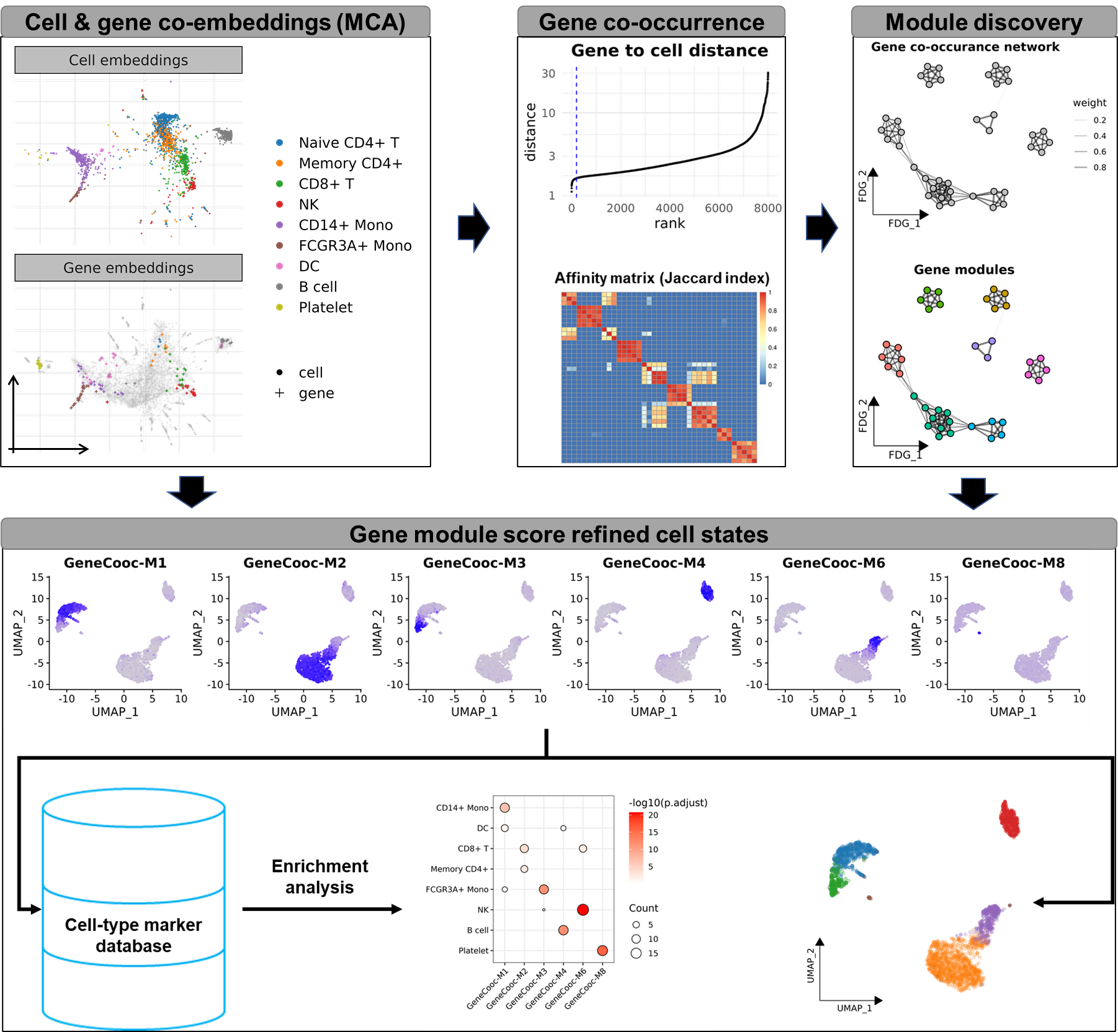
GeneCooc is an ultra-fast R package for gene co-expression module
discovery facilitates cell state identification.
You can install the development version of GeneCooc from
GitHub with:
# install.packages("devtools")
# install CelliD (Seurat V4)
devtools::install_github("RausellLab/CelliD", ref="e306b43")
# install GeneCooc
devtools::install_github("JarningGau/GeneCooc")Note: GeneCooc is compitable with Seurat V4, we did not test the codes
on Seurat V5.
- Calculate the gene to cell distance and rank the genes by this distance for each cell.
seu <- CalGeneRankings(seu, min.expr.cells = 100)- Calculate the gene affinity matrix. The gene affinity is measured by co-occurance ratio.
seu <- CalAffinityMatrix(seu, K = 200, min.freq = 10)- Find modules.
The major modules are divided by louvain cluster on gene-gene coexpression graph defined by gene affinity matrix. Then minor modules are divided using the dynamic tree cut on a hierarchical tree for each major module.
seu <- FindModules(seu)- Trim the minor modules by archytype analysis.
seu <- TrimModules(seu)- Merge the similary minor modules automaticall。
seu <- AutoMergeModules(seu)- Scoring each gene module.
seu <- CalModuleScore(seu)