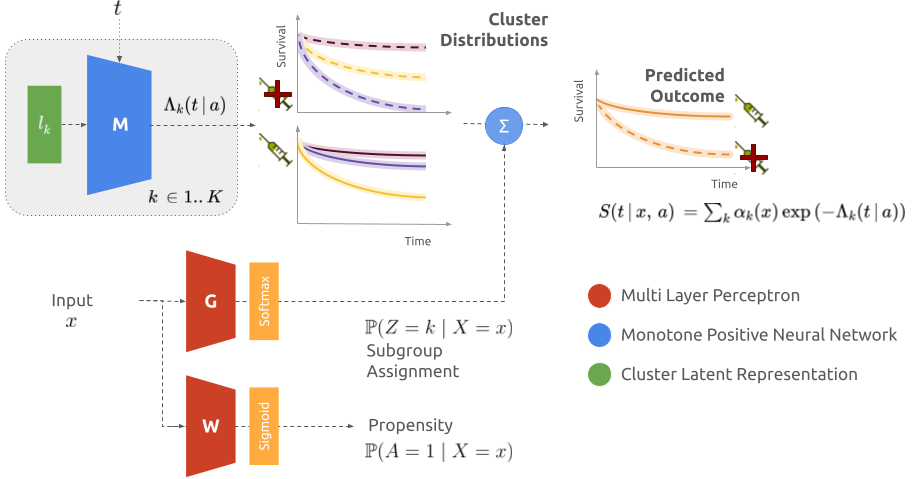
This repository allows to use the model and reproduce the results introduced in Identifying treatment response subgroups in observational time-to-event data. This models aim to uncover subgroups of patients with different treatment effects. Each patient is assigned to a subgroup characterised by two neural networks modelling survival under treatment and control regimes.
The model consists of three neural networks for estimating the survival distributions: M models the cumulative incidence function under both treatment regimes given a latent representation for each cluster, G assigns each patient to the different subgroups, and W computes the probability to receive treatment to adjust the likelihood for observational study.
To use the model, one needs to execute:
from cnsc import CausalNeuralSurvivalClustering
model = CausalNeuralSurvivalClustering()
model.fit(x, t, e, a)
model.predict_risk(x, risk = 1)With x the covariates, t the time of observed events, e the associated cause (0 if censored, 1 if the outcome of interest) and a the binary treatment.
A full example with analysis is provided in examples/Causal Neural Survival Clustering on METABRIC Dataset.ipynb using a publicly available dataset for reproducibility. Note that this dataset does not meet the assumptions necessary to estimate treatment effect, and should consequently only be used as a tutorial on how to use the model.
To reproduce the paper's results:
- Clone the repository with dependencies:
git clone git@github.com:Jeanselme/CausalNeuralSurvivalClustering.git --recursive. - Create a conda environment with all necessary libraries
pycox,lifelines,pysurvival. - Add path
export PYTHONPATH="$PWD:$PWD". - Run
examples/experiment_cnsc.py SEER. - Analysis using
examples/Analysis CNSC.ipynb.
Adding a new method consists in adding a child to Experiment in experiment.py with functions to compute the nll and fit the model.
Then, add the method in examples/experiment_cnsc.py and follow the previous point.
We followed the same architecture than the DeepSurvivalMachines repository with the model in cnsc/ - only the api should be used to test the model. Examples are provided in examples/.
git clone git@github.com:Jeanselme/CausalNeuralSurvivalClustering.git
The model relies on pytorch >= 2.0, numpy and tqdm.
To run the set of experiments auton_survival, pycox, lifelines, pysurvival are necessary.