ReadBouncer: Precise and Scalable Adaptive Sampling for Nanopore Sequencing
Jens-Uwe Ulrich, Ahmad Lutfi, Kilian Rutzen, Bernhard Y. Renard
Bioinformatics, Volume 38, Issue Supplement_1, July 2022, Pages i153–i160, https://doi.org/10.1093/bioinformatics/btac223
ReadBouncer is a nanopore adaptive sampling tool for Windows and Linux (x64 or ARM64) that uses Interleaved Bloom Filters for live classification of nanopore reads, basecalled with either Guppy(GPU mode) or DeepNano-blitz(CPU mode). The Toolkit uses Oxford Nanopore's Read Until functionality to unblock reads that match to a given reference sequence database. The database is indexed as Interleaved Bloom Filter for fast classification.
- In a first step the reference sequences are used to build an Interleaved Bloom Filter using the SeqAn library
- Interleaved Bloom Filters can be used as depletion or enrichment target
- After Starting a sequencing run, ReadBouncer receives raw current signals from the sequencer via ONT's MinKNOW API using Google's remote procedure calls (gRPC)
- Signals are basecalled in real-time with DeepNano-blitz or Guppy
- Basecalled reads are matched to the reference sequences in the Interleaved Bloom Filters and unblock/discard messages are sent back to the sequencer according to the read classification
The easiest way is to download the provided installer files for Windows, Linux x86_64 or Linux arm64 and simply click through the installation process.
ReadBouncer has the following dependencies that should be installed before compiling the source code
- CMake for building
- C++ Compiler Currently we support MSVC (on Windows) and GCC-8 or later (on Linux)
- Perl
- Go
- NASM Compiler
- NSIS Needed for creation of installer executables
- UUID On Linux machines,
uuidanduuid-devneed to be installed (e.g.sudo apt install uuid uuid-dev) - TAR On Windows, make sure tar is installed on the system
Then just need to clone the repository to your computer, create a build directory within cloned directory and let cmake do the work for you
Compiling the source code yourself on Windows just requires the following command line calls
git clone https://github.com/JensUweUlrich/ReadBouncer.git
cd .\ReadBouncer\
mkdir build
cd .\build\
cmake.exe ..\src
cmake.exe --build . --config Release
cmake.exe --build . --config Release --target package
The last step creates the ReadBouncer-1.2.2-win64.exe within the build directory, which is a simple installer for Windows that leads you through the installation process.
Compiling the source code Linux works similar to Windows. Just open a terminal, change to your working directory of choice and use the following commands
git clone https://github.com/JensUweUlrich/ReadBouncer.git
cd ReadBouncer
mkdir build
cd build
cmake ../src
cmake --build . --config Release
cmake --build . --config Release --target package
The last step creates the ReadBouncer-1.2.2-Linux.sh within the build directory, which is a simple command line installer for Linux that leads you through the installation process. You can also skip the last cmake step and just call sudo make install, which installs ReadBouncer in your /usr/local/ directory.
ReadBouncer can simply be called from the command line by providing a TOML configuration file.
full\path\to\ReadBouncer\root\directory\bin\ReadBouncer.exe --config full\path\to\ReadBouncer\config.toml
All parameters, input and output files and the usage are specified within the configuration file. First, there are three different modes for using ReadBouncer (build, target and classify). The build usage is specified when the goal is to create an Interleaved Bloom Filter index file. The classify usage is specified when only a read classification with based on ReadBouncer's IBF-based classifier shall be tested, and the The target usage is specified when ReadBouncer shall be used in an adaptive sampling experiment. You can also specify an output and a log directory.
usage = "build", "target", "classify", "test" #atm only one of those
output_directory = 'path/to/write/output/files/to' #all generated output files will be stored here
log_directory = 'path/to/write/log/files/to' #all generated log files will be stored here
Before using ReadBouncer for adaptive sampling, you may want to create the reference database(s) for target and/or depletion reference sequences with the usage build using the config.toml file. In this step you have to provide the reference sequence(s) as a comma-separated list of FASTA files (target/depletion files), the fragment size and the size of the kmers used to build the IBF. The resulting Interleaved Bloom Filter files will be stored in the given output directory.
usage = "build"
output_directory = 'path/to/write/output/files/to' #all generated output files will be stored here
log_directory = 'path/to/write/log/files/to' #all generated log files will be stored here
[IBF]
kmer_size = X #(unsigned integer; default: 13)
fragment_size = X #(unsigned integer; default: 100000)
threads = X #(unsigned integer; default: 3)
target_files = ['xxx.fasta', 'xxx.fasta']
deplete_files = ['xxx.fasta', 'xxx.fasta']
kmer_size
For every fragment we compute all k-mers of this size, calculate hash values for the k-mers and add those to the Interleaved Bloom Filter like described by Dadi et. al., 2018. For adaptive sampling experiments using DeepNano for basecalling, we expect the sequencing error rates to be larger than 10%. Here, we recommend using a kmer_size of 13 and fragment_size of 100,000, whereas for experiments with Guppy live-basecalling, a kmer_size of 15 and fragment_size of 200,000 may yield best results.
fragment_size
The reference sequence is fragmented in subsequences of this length, where every fragment is stored in a separate bin of the Interleaved Bloom Filter. Fragments overlap by 1,500 nucleotides because ReadBouncer only tries to classify and unblock a nanopore read until the first 1,500 nucleotides have been sequenced. The fragmentation leads to better classification specificity when using smaller kmer_size values.
If you like to test ReadBouncer's read classification with a set of Nanopore reads, you can use the classify usage. You only have to provide one or more Interleaved Bloom Filter (IBF) or FASTA files (automatically create ibf file for every given fasta file) and some reads as FASTA or FASTQ file. When providing already created IBF files, make sure that the files are located in the given output directory. When providing an IBF file as depletion-file ReadBouncer takes the prefix from each read and maps it against each bin of the depletion-IBF. If the number of k-mers that are shared between the prefix and at least one bin of the IBF exceeds a certain threshold, the read is classified as match. The threshold is calculated as described in the publication. If you provide a target IBF file as well, ReadBouncer will not classify reads if the prefix matches the depletion IBF but not the target IBF. Providing only target_files will lead ReadBouncer to assign each read to the best matching target filter file. Result files of the classification can also be found in the output directory.
usage = "classify"
output_directory = 'path/to/write/output/files/to' #all generated output files will be stored here
log_directory = 'path/to/write/log/files/to' #all generated log files will be stored here
[IBF]
kmer_size = X #(unsigned integer; default: 13)
fragment_size = X #(unsigned integer; default: 100000)
threads = X #(unsigned integer; default: 3)
target_files = ['xxx.fasta', 'xxx.fasta'] or ['xxx.ibf', 'xxx.ibf']
deplete_files = ['xxx.fasta', 'xxx.fasta'] or ['xxx.ibf', 'xxx.ibf']
read_files = ['xxx.fasta', 'xxx.fasta'] or ['xxx.fastq', 'xxx.fastq']
exp_seq_error_rate = 0.1 #(unsigned float between 0 and 1; default: 0.1)
chunk_length = X #(unsigned integer; default: 250)
max_chunks = X #(unsigned integer; default: 5)
chunk_length and max_chunks
These parameters decide about the number of nucleotides from the beginning of each read that is used for classification. ReadBouncer tries to classify reads in in chunk-wise manner, when using e.g. 2 chunks with 360 bp length, ReadBouncer takes the first 360 bp of the read for classification and will add the subsequent 360 bp of that read for another classification try if the first try did not succeed.
deplete_files and target_files
The deplete_files contain the reference sequences for reads we would like to reject. Providing only deplete_files will lead ReadBouncer to classify given reads as rejected if their prefixes match against at least one of the given files. If we also have a priori knowledge about target reference sequences, we can provide ReadBouncer with target_files and reads are assigned to the best matching file.
exp_seq_error_rate
For more accurate classification of reads we are calculating the expected number of mutated k-mers for each read prefix based on the expected sequencing error rate. Than a confidence interval for the mutated k-mers is calculated as described by Blanca et. al., 2021 and the minimum number of matching k-mers is calculated based on the upper bound of the confidence interval. The significance level of the confidence interval is set to 95% by default.
fragment_size and kmer_size
Both parameters are only required if the given target or deplete files are in FASTA format. In this case we need to build the Interleaved Bloom Filters directly. They are ignored if already built IBF files are provided.
When nanopore reads are not of interest, these reads can be rejected from the pore without sequencing the whole read. Therefore raw current signals are requested from ONT's MinKNOW software, basecalled and matched against one or more Interleaved Bloom Filter of a give reference sequence set. Depending on the given combinations of deplete and target files, ReadBouncer's adaptive sampling behaves different. a) If only deplete_files are provided, every read that matches with at least one of the given file is rejected from the corresponding pore by sending an unblock message to MinKNOW for that read. b) If only target_files are provided, all reads that do not match to at least one target file will be unblocked. c) If both target_files and deplete_files are provided, only reads that match to at least one of the deplete_files, but to none of the target_files are unblocked. This approach can be used to ensure high specificity if sequences in depletion and target reference databases have certain regions of high similarity.
usage = "target"
output_directory = 'path/to/write/output/files/to' #all generated output files will be stored here
log_directory = 'path/to/write/log/files/to' #all generated log files will be stored here
[IBF]
kmer_size = X #(unsigned integer; default: 13)
fragment_size = X #(unsigned integer; default: 100000)
target_files = ['xxx.fasta', 'xxx.fasta'] or ['xxx.ibf', 'xxx.ibf']
deplete_files = ['xxx.fasta', 'xxx.fasta'] or ['xxx.ibf', 'xxx.ibf']
exp_seq_error_rate = 0.1 #(unsigned float between 0 and 1; default: 0.1)
threads = X #(unsigned integer; default: 3) classification threads
[MinKNOW]
host = "xxx.xxx.xxx" #(ip address or name of the computer hosting MinKNOW; default: 127.0.0.1)
port = "XXXX" #(port number used fo grpc communication by MinKNOW instance; default: 9502)
flowcell = "XXX" #(name of the flowcell used)
token_path = 'path/to/minknow-auth-token.json' #path to authentication token file (remote file)
[Basecaller]
caller = "DeepNano" or "Guppy" #(default: DeepNano)
host = "xxx.xxx.xxx" #(ip address or name of the computer hosting Guppy Basecall Server, or ipc path, e.g. ipc:///tmp/.guppy; default: 127.0.0.1)
port = X #(port number on which the basecall server is running on the host; default: 5555)
threads = X #(unsigned integer; default 3) basecall threads; only required for DeepNano base-calling
config = "config_name" #(only needed for Guppy; name of the Guppy basecalling config, e.g. "dna_r9.4.1_450bps_fast")
[IBF] fragment_size and kmer_size
Both parameters are only required if the given target or deplete files are in FASTA format. In this case we need to build the Interleaved Bloom Filters directly. They are ignored if already built IBF files are provided.
[IBF] deplete_files and target_files
The deplete_files (as IBF or FASTA file) contain the reference sequences we want to deplete. In other words, nanopore reads that match against these references shall be rejected from the pore. It is possible to directly provide the files in FASTA format and assign the parameters to build the IBF. ReadBouncer builds automatically an IBF for each provided FASTA file using fragment_size and kmer_size. If we also have a priori knowledge about potential organisms or genomic regions of interest in our sample that should not be rejected, it is also possible to build an IBF of the reference sequences of those organisms or genomic regions and provide ReadBouncer this information as target IBF file. This can improve the specificity of the classification.
[IBF] exp_seq_error_rate
For more accurate classification of reads we are calculating the expected number of mutated k-mers for each read prefix based on the expected sequencing error rate. Than a confidence interval for the mutated k-mers is calculated as described by Blanca et. al., 2021 and the minimum number of matching k-mers is calculated based on the upper bound of the confidence interval. The significance level of the confidence interval is set to 95% by default.
[MinKNOW] host and port
This is the IP adress and the TCP/IP port on which the MinKNOW software is hosted. ReadBouncer will exchange data with the MinKNOW software via this communication channel. ReadBouncer automatically tests if a valid connection can be established before starting the sequencing run.
[MinKNOW] flowcell
This is the name of the FlowCell for which we want apply adaptive sampling. (Can be found in MinKNOW GUI)
[MinKNOW] token_path
The path to the token file for the remote connection. ReadBouncer finds the path automatically in local connection mode. For remote connection, this file is located at /tmp/minknow-auth-token.json on the machine running minknow and has to be copied to the same device running ReadBouncer. Please note that you have to enable guest mode on the sequencer in the minknow user config file by changing guest_rpc_enabled to enabled.
[Basecaller] caller
Basecaller used during adaptive sampling. For CPU base-calling use "DeepNano", use "Guppy" for GPU base-calling otherwise. Please note that you need to start the Guppy basecall server on a host machine with powerful GPUs that can keep up with the sequencing speed. ReadBouncer will connect to the server via its integrated the guppy basecall client. We recommend read Miles Benton's great github repository on setting up adaptive sampling with NVIDIA AGX/NX. We further recommend testing adaptive sampling with a playback run before starting a real experiment.
[Basecaller] host
IP address of guppy basecall server or path to IPC channel. Only required for Guppy live base-calling. Please use Guppy only in Fast mode in order to keep up with the sequencing pace. Since version 6, Guppy by default uses IPC as communication channel, then you need to provide the ipc path (e.g. ipc:///tmp/.guppy) as host. However this only works if ReadBouncer and Guppy run on the same machine. If both tools run on different machines, you need to start Guppy with options ´--use_tcp´ and ´--allow_non_local´
[Basecaller] port
TCP/IP port of guppy basecall server. Only required for Guppy live base-calling. By default, MinKNOW starts a Guppy basecall server on port 5555. If you start a different instance on another port, you have to provide the port here.
[Basecaller] threads
Number of threads used for base calling. This parameter only has an effect if CPU basecalling with DeepNano-blitz is used.
[Basecaller] config
Name of Guppy's baecalling config used by the guppy_basecall_server. Not required when using DeepNano as basecaller.
Sometimes it can be useful to find all reads of an organism in a set of reads that were already sequenced without aligning the sequences. ReadBouncer offers this functionality by using the classify subcommand. The following steps describe how to classify all reads from a Zymo Mock Community to their corresponding reference genome.
- Download the reference sequences of the Zymo Mock Community from here and store it in your working directory.
- Create a configuration file similar to the following one, providing all necessary parameters, file and directory paths in the config.toml file.
usage = "classify"
output_directory = 'full/path/to/ReadBouncer/output_dir/'
log_directory = 'full/path/to/ReadBouncer/log/files'
[IBF]
kmer_size = 13
fragment_size = 100000
threads = 3
target_files = ['path/to/reference/file/Bacillus_subtilis_complete_genome.fasta', 'path/to/reference/file/Enterococcus_faecalis_complete_genome.fasta', 'path/to/reference/file/Escherichia_coli_complete_genome.fasta']
deplete_files = ['path/to/reference/file/Saccharomyces_cerevisiae_draft_genome.fasta']
read_files = ['path/to/read/file/SampleZMCDataSet.fasta']
In this example, we would try to find all reads that match to B.subtilis, E.faecalis and E.coli, but not to S.cerevisiae. You can easily add the other fasta files as well. Now we can start ReadBouncer from the command line using the config.toml file. For testing purposes, you can download a small set of sequenced Zymo Mock community nanopore reads that were basecalled with DeepNano (sample read set)
full\path\to\ReadBouncer\root\directory\bin\ReadBouncer.exe --config full\path\to\ReadBouncer\config.toml
Finally, you will get some stats printed to the command line, as the one below:
------------------------------- Final Results -------------------------------
Number of classified reads : 47874
Number of of too short reads (len < 250) : 0
Number of all reads : 100000
Bacillus_subtilis_complete_genome : 14409
Enterococcus_faecalis_complete_genome : 13553
Escherichia_coli_complete_genome : 19912
Average Processing Time Read Classification : 0.00197617
-----------------------------------------------------------------------------------
Resulting FASTA files and index files can be found in the provided output directory.
Before using ReadBouncer in a real experiment, we recommend running a playback experiment testing also basecalling and classification performance.
-
Download an open access bulk FAST5 file from here. This file is 21Gb so make sure you have plenty of space.
-
To configure a run for playback, you need to find and edit a sequencing TOML file. These are typically located in
C:\Program Files\OxfordNanopore\MinKNOW\conf\package\sequencingon Windows or/opt/ont/minknow/conf/package/sequencingon Linux. Edit a file such as sequencing_MIN106_DNA.toml and under the entry[custom_settings]add a field:simulation = "/full/path/to/your_bulk.FAST5" -
Since MinKNOW 5.1, we can only create a simulated device by proving the MinKNOW service executable the corresponding parameter. On Linux this can be done by editing the file
/lib/systemd/system/minknow.service:
[Service]
ExecStart=/opt/ont/minknow/bin/mk_manager_svc --simulated-integrated-devices 1
Common error:Invalid local-auth tokencan be solved byexport MINKNOW_API_USE_LOCAL_TOKEN=1 -
Then you need to reload the daemon via
sudo systemctl daemon-reloadand restart the MinKNOW service viasudo service minknow restart(In some cases you need to reboot your operating system). Your MinKNOW instance will now show a simulated device namedMS00000that will playback the bulkfile rather than live sequencing. -
Open a Windows Power Shell (or terminal) and go to your working directory where ReadBouncer result files shall be stored. Then provide the necessary parameters in the config.toml file for the test. ReadBouncer will test the conncetion to MinKNOW automatically and tell you when to start the experiment from within MinKNOW. You can download two fasta files that include reference sequences for human chromosomes 21 & 22 as well as the remaining chromsome reference sequences from the Telomere-to-telomere consortium. You should unzip them and use the following example toml file to test an experiment that targets the sequencing of reads that belong to chromosomes 21 & 22. All other reads shall be rejected/unblocked.
usage = "target"
output_directory = 'full/path/to/ReadBouncer/output_dir/'
log_directory = 'full/path/to/ReadBouncer/'
[IBF]
kmer_size = 13
fragment_size = 100000
threads = 3
target_files = ['path/to/reference/sequence/chm13.v1.1_chr21_22.fasta']
deplete_files = ['path/to/reference/sequence/chm13.v1.1_chr1-20_XM.fasta']
exp_seq_error_rate = 0.1
[MinKNOW]
host = "localhost"
port = "9502"
flowcell = "MS00000"
token_path = "test/tmp/minknow-auth-token.json" #path to authentication token file (if not localhost)
[Basecaller]
caller = "DeepNano"
threads = 3
Using command line:
full\path\to\ReadBouncer\root\directory\bin\ReadBouncer.exe --config full\path\to\ReadBouncer\config.toml
- Now ReadBouncer will construct Interleaved Bloom Filters for both reference files. After finishing the construction, IBF files are stored in the output directory for for further use. ReadBouncer will tell you if the connection to the MinKNOW host has been successfully established and that you can start sequencing now.
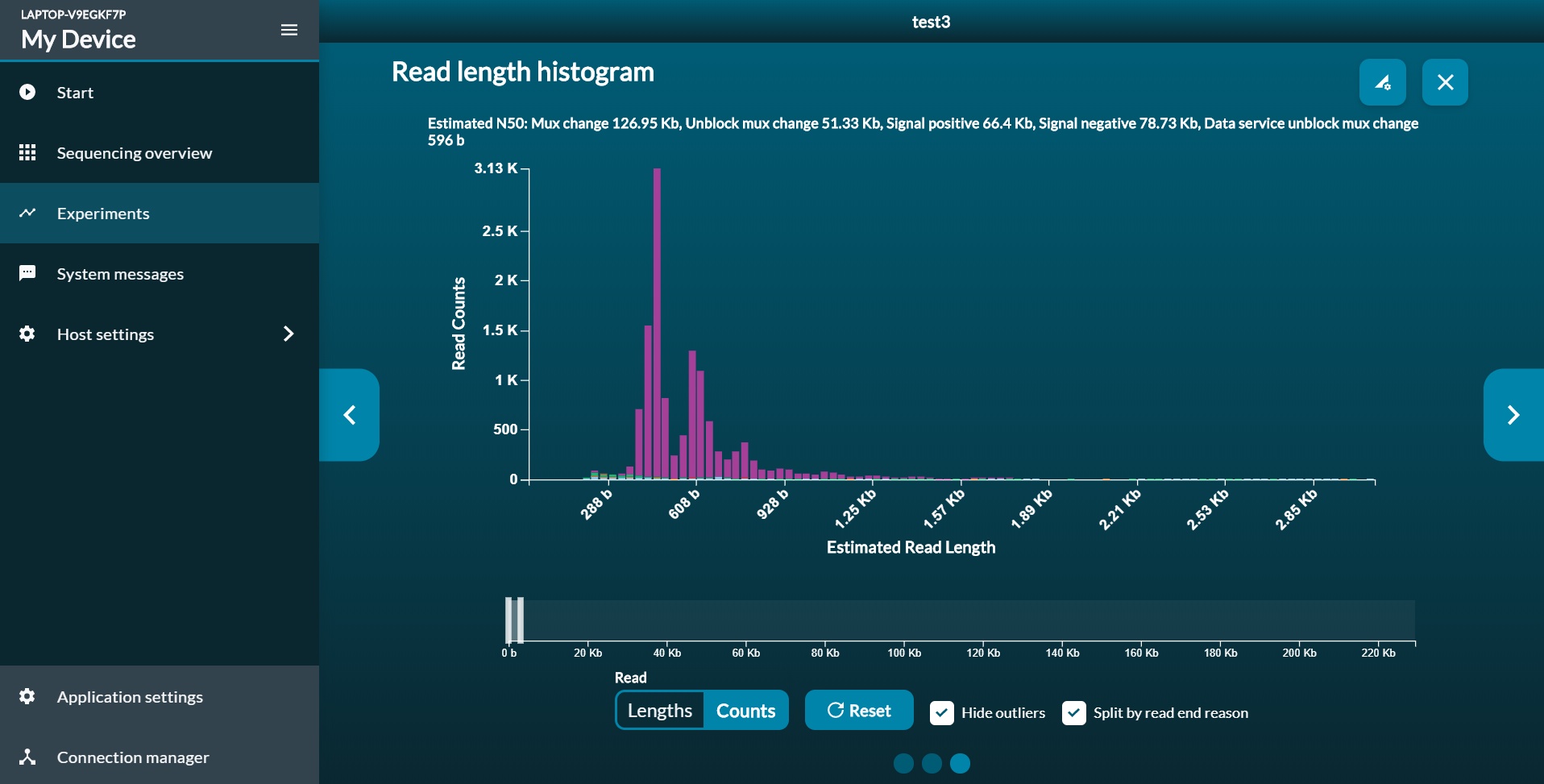
- Start a sequencing run on the simulated device from within the MinKNOW-GUI. Open the read length histogram after 15 minutes and have a look at the read counts plot. When you zoom into the region for reads up to 5kb length, you should see a plot like this:
- After the run finished, ReadBouncer will provide you with some statistics about the number of classified (unblocked) and unclassified reads, which will be sequenced until the end. You will also see average overall processing times as well as for basecalling and classification. You should aim for overall processing times for classified reads below one second. The average processing time for basecalling and classification should be below 0.01 seconds. Otherwise you will experience bigger lengths of unblocked reads.
- ReadBouncer is highly dependent on basecallers and their accuracy. Since the base-calling accuracy of ONT's Guppy (in fast mode) is much higher than that of DeepNano-blitz, we highly recommend to use Guppy if GPUs are available. If you are not sure whether your GPU is powerful enough to perform live-basecalling, you can have a look at Miles Benton's great github repository, where he summarizes all his experiences with Adaptive Sampling on different GPUs.
- At the moment, CPU base-calling with DeepNano-blitz is not supported on ARM64.