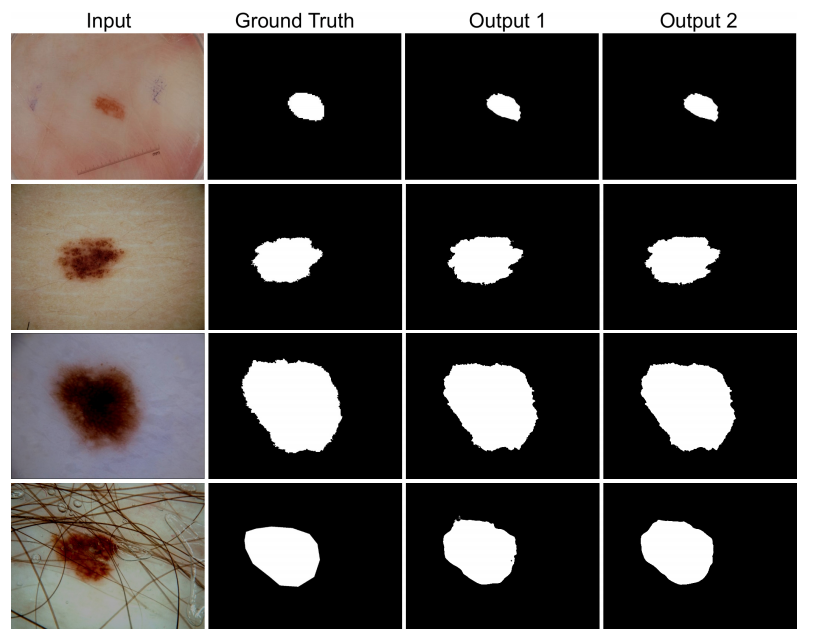
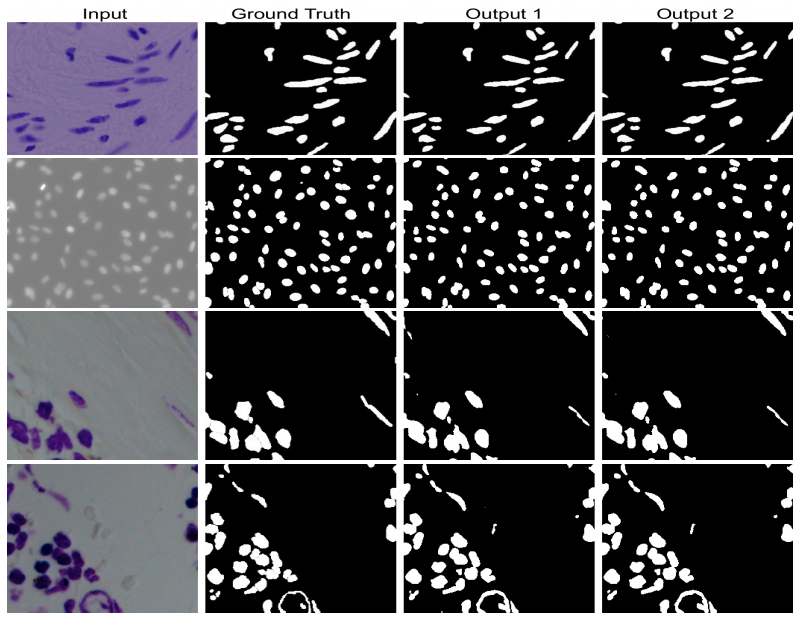
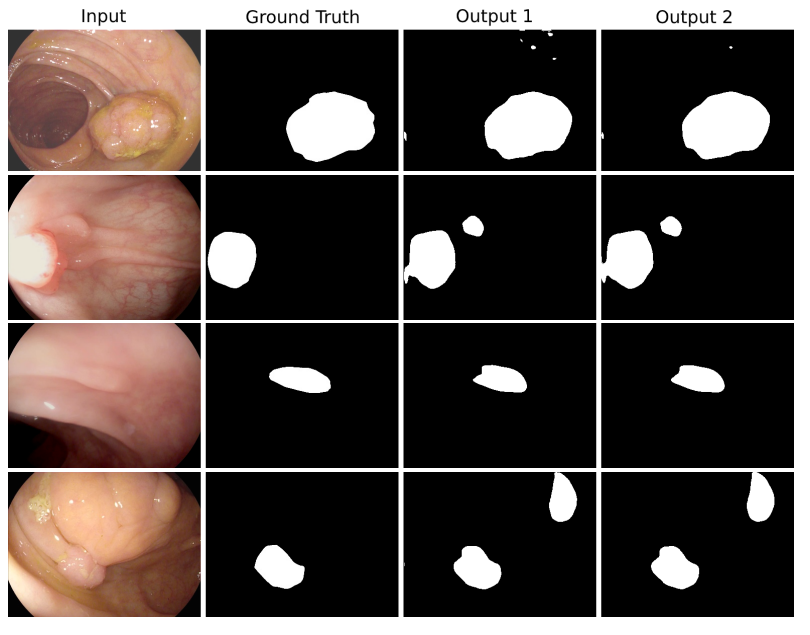
DoubleU-Net is an enhanced U-Net architecture that integrates the power of pre-trained models and multi-scale feature extraction capabilities. This repository is a comprehensive guide to understanding and applying DoubleU-Net for various medical segmentation tasks.
Enhanced U-Net Structure: DoubleU-Net comprises two U-Net structures combined innovatively. This design is intended to capture richer contextual information and deliver better segmentation performance.
Improved Feature Extraction: DoubleU-Net employs a VGG19 architecture (pre-trained on ImageNet) as the encoder for the first U-Net. This allows the model to harness the feature extraction capability of VGG19, which is robust for many visual tasks.
ASPP (Atrous Spatial Pyramid Pooling) Module: The paper introduces an ASPP module between the two U-Net structures. ASPP has been instrumental in semantic segmentation tasks because it captures multi-scale information. By incorporating it into the DoubleU-Net, the model is better equipped to handle objects of varying sizes in medical images.
Performance: The proposed DoubleU-Net was tested on several medical image segmentation datasets and outperformed other state-of-the-art models in many cases.
Encoder: Start with a pre-trained VGG19 encoder. This will form the encoder part of the first U-Net.
U-Net Architecture: Implement the decoder part for the first U-Net, ensuring to have skip connections from the encoder. Follow this with the ASPP module.
Second U-Net: The ASPP module output serves as the second U-Net's encoder. Implement this U-Net ensuring it captures multi-scale features effectively.
Loss Function: Choose an appropriate loss function Depending on the dataset and segmentation task. Dice loss or a combination of Dice and cross-entropy loss is commonly used for medical image segmentation.
Data Augmentation: Medical datasets can often be small. Augmenting the data can help in training the model better. Consider rotations, flips, and elastic deformations common for medical images.
Evaluation Metrics: Use appropriate evaluation metrics like the Dice coefficient, Jaccard index, or others suitable for the specific segmentation task.
- Pre-trained Model Integration: Utilizing established models for better encoders.
- Multi-scale Segmentation: Addressing varying object sizes with the ASPP module.
- Generalizability: Reducing dataset-specific architecture tweaks.
- Efficient Learning: Superior results even with limited training data.
- Reference Model: Benchmarking and comparison with other segmentation architectures.
- Retinal Image Segmentation: Segment vessels and structures in fundus images.
- Lung Field Segmentation: Outline lung fields in chest X-rays.
- Tumor Detection: Segment tumors in MRI or CT slices.
- Skin Lesion Isolation: Identify lesions or melanomas in dermoscopic images.
- Bone Segmentation: Highlight fractures or abnormalities in X-rays.
- Liver Segmentation: Differentiate liver in abdominal CT scans.
- Brain Structure Segmentation: Identify cerebellum, cortex, or ventricles in MRI.
- Blood Clot Detection: Locate clots or plaques in vascular ultrasound images.
- Heart Chamber Segmentation: Distinguish heart chambers in echocardiograms.
- Kidney and Tumor Segmentation: Differentiate kidney tissues and tumors in medical scans.
Please cite our paper if you find the work useful:
@INPROCEEDINGS{9183321,
author={D. {Jha} and M. A. {Riegler} and D. {Johansen} and P. {Halvorsen} and H. D. {Johansen}},
booktitle={2020 IEEE 33rd International Symposium on Computer-Based Medical Systems (CBMS)},
title={DoubleU-Net: A Deep Convolutional Neural Network for Medical Image Segmentation},
year={2020},
pages={558-564}}
please contact debesh.jha@northwestern.edu for any further questions.