This repo contains codes and data for priority program, which is under review of One Earth of Cell Press. I need to rerun AOH to generate correct species data to do prioritization.
I have downloaded species range data from IUCN Red List Resources, and get the file(this file is not uploaded here, see workstation209 directory) AMPHIBIANS.zip. Trying to generate AOH using AmphibianAOH.R code.
All goes well. Now we have 8861 rows in amphi_range_data and 6989 rows in amphi_info_data. I will check the difference between them after all AOHs have been generated. Estimated time is 2d. 2 days passed and all goes well. We have generated 6771 amphibian tifs. The difference happens from 8861 to 6989 is because one species have different sources. The difference happens from 6989 to 6771 is because some habitat code in not included in Lumbierres dataset.
The final amphibian list data amphi_spp_list.csv and amphibian aoh raster tifs are stored in workstation 209, amphi_spp_list.csv also uploaded. The processing rds files are in the same directory with amphi_spp_list.csv, but not uploaded here.
I downloaded species range data from IUCN Red List Resources, and the hydrobasin one only includes a csv file, without any spatial information, so I dropped it. The file REPTILES.zip is also in workstation209 directory. Code is ReptileAOH.R.
The workstation's memory only supports processing one taxa at one time. Running reptiles' aoh now.
A lot of problems arise.
-
There are some species whose AOH cannot be generated, because the create_spp_aoh() step need more memory than our workstation's RAM(128G). This problem has been fixed after the update of aoh package.
-
We cannot generate reptile's spp_info_data, because this step still needs much memory. So we are trying to split the reptile taxa into a few groups, and process each group one by one. Unfortunately, it still failed to work when there is only one species each group. It's confusing and I don't know what to do.
I use the code MammalAOH_narrow.R to generate the AOH of error mammals. The error list is generated in April, 2022, using geometry attribute in MAMMALS.rds sent from Jeff. Hope this will work.
Unfortunately, this ended up with blowing up the memory again. So I just try to run the narrow mammal species one by one using MammalAOH_OnebyOne.R. This works! Output species list in mammal_spp_list[ReAOH/mammal_spp_list.csv]
Use the code ReptileAOH_Narrow_OnebyOne.R to generate narrow reptile species' AOH, it works well. Output species list in reptl_spp_list.csv
Jeff finished the narrow species list of bird taxa, all files in aoh_redo_birds, I cleaned up the bird_spp_list and modified file path according to results.rda.
Processing R script is mammal_5km.R, amphibian_5km.R, reptile_5km.R and bird_5km_useFunction.R.
All done! SPList for amphibian, reptile and mammal are generated in the loop, now generate independently for birds: bird_5km_splist.R
Done!
I use former method to generate riij matrix for four taxa. Codes are rij_amphibian,rij_reptile,rij_bird,rij_mammal. Generated rij matrices are rij_amphibian,rij_reptile,rij_bird,rij_mammal.
I downloaded the carbon raster tif from open data of the paper Mapping the irrecoverable carbon in Earth’s ecosystems. We use Vulnerable Carbon Total 2018 version.
The crs of raster is WGS84, and the unit is degree. Overall pattern looks like this: 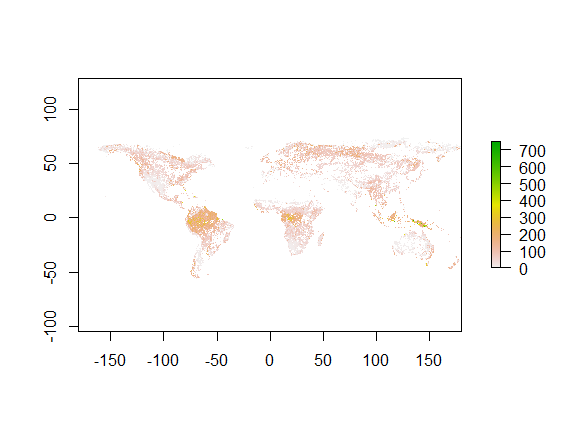
I will use ArcGIS to reproject it to Mollweide, and then resample to 5 km. Lastly extract with grid_cell points. The final output is 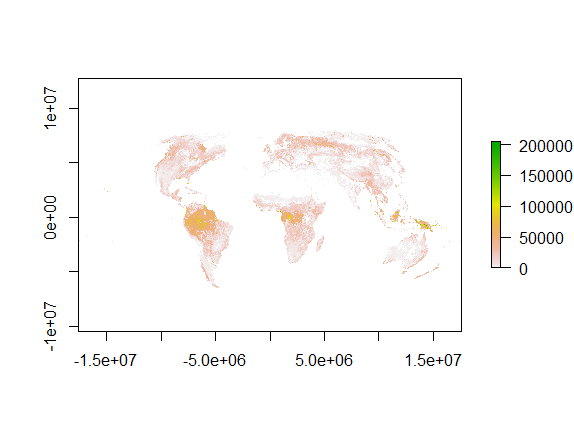
I use the same settings as last time: no lock out urban, delete Mergus merganser_2 as its AOH too sparse to achieve targets. Codes are Country scenario 30% area, Country scenario 50% area, Global scenario 30% area, Global scenario 50% area. Again I use the function to count how many species have reached targets. I use previous code to calculate how much area have been protected and selected in each country and ecoregion.
I use add_shuffle_portfolio to reduce the stripes in country scenario outputs, following Jeff's suggestions. Much better.
This time , Global scenario 50% area 0 weight used up to more tha 90% of the budget, far more than last time (~40%), causing diffrent pattern. I checked the species aoh and all is fine. So I am guessing if prioritizr doesn't mind budget usage as long as it's under the threshold. As to improve computing speed, use much budget at first may help? Now I am generating portfolios for glo50 0weight to see if budget usage varies a lot.