| title | output | ||||
|---|---|---|---|---|---|
Scripts for exploring regulatory potential of predefined regions |
|
Here is a set of scripts for the for exploring regulatory potential of predefined regions.
usage: makeWindows.py [-h] -A ANNOTATION_FILE [-HWP WINDOW_PROMOTER] [-HWE WINDOW_ENHANCER] [-BN BIN_NUMBERS] [-C CUT_LENGTH] [-L MIN_LENGTH] -O OUTPUT_TAG
optional arguments:
-h, --help show this help message and exit
-A ANNOTATION_FILE, --annotation_file ANNOTATION_FILE
Gene annotation file.
-HWP WINDOW_PROMOTER, --window_promoter WINDOW_PROMOTER
Half window size for promoters [100].
-HWE WINDOW_ENHANCER, --window_enhancer WINDOW_ENHANCER
Half window size for enhancers [10000].
-BN BIN_NUMBERS, --bin_numbers BIN_NUMBERS
Number of bins for gene bodies [20].
-C CUT_LENGTH, --cut_length CUT_LENGTH
Numober of bases to cut from gene bodies [1000].
-L MIN_LENGTH, --min_length MIN_LENGTH
Minimal length of gene body [1000].
-O OUTPUT_TAG, --output_tag OUTPUT_TAG
Prefix for output files.
The ANNOTATION_FILE is provided for human genome hg19 in the out folder. To run the script with default setting:
python makeWindows.py -A GeneAnnotation_Hg19.tsv -O hg19
The resulting file 'hg19_windows.bed' contains genomic intervals regarding gene structures for each gene:
chr1 68990 69190 ENSG00000186092_OR4F5_P_0
chr1 68790 68990 ENSG00000186092_OR4F5_PU_1
chr1 68590 68790 ENSG00000186092_OR4F5_PU_2
chr1 68390 68590 ENSG00000186092_OR4F5_PU_3
chr1 68190 68390 ENSG00000186092_OR4F5_PU_4
chr1 67990 68190 ENSG00000186092_OR4F5_PU_5
chr1 67790 67990 ENSG00000186092_OR4F5_PU_6
chr1 67590 67790 ENSG00000186092_OR4F5_PU_7
chr1 67390 67590 ENSG00000186092_OR4F5_PU_8
chr1 67190 67390 ENSG00000186092_OR4F5_PU_9
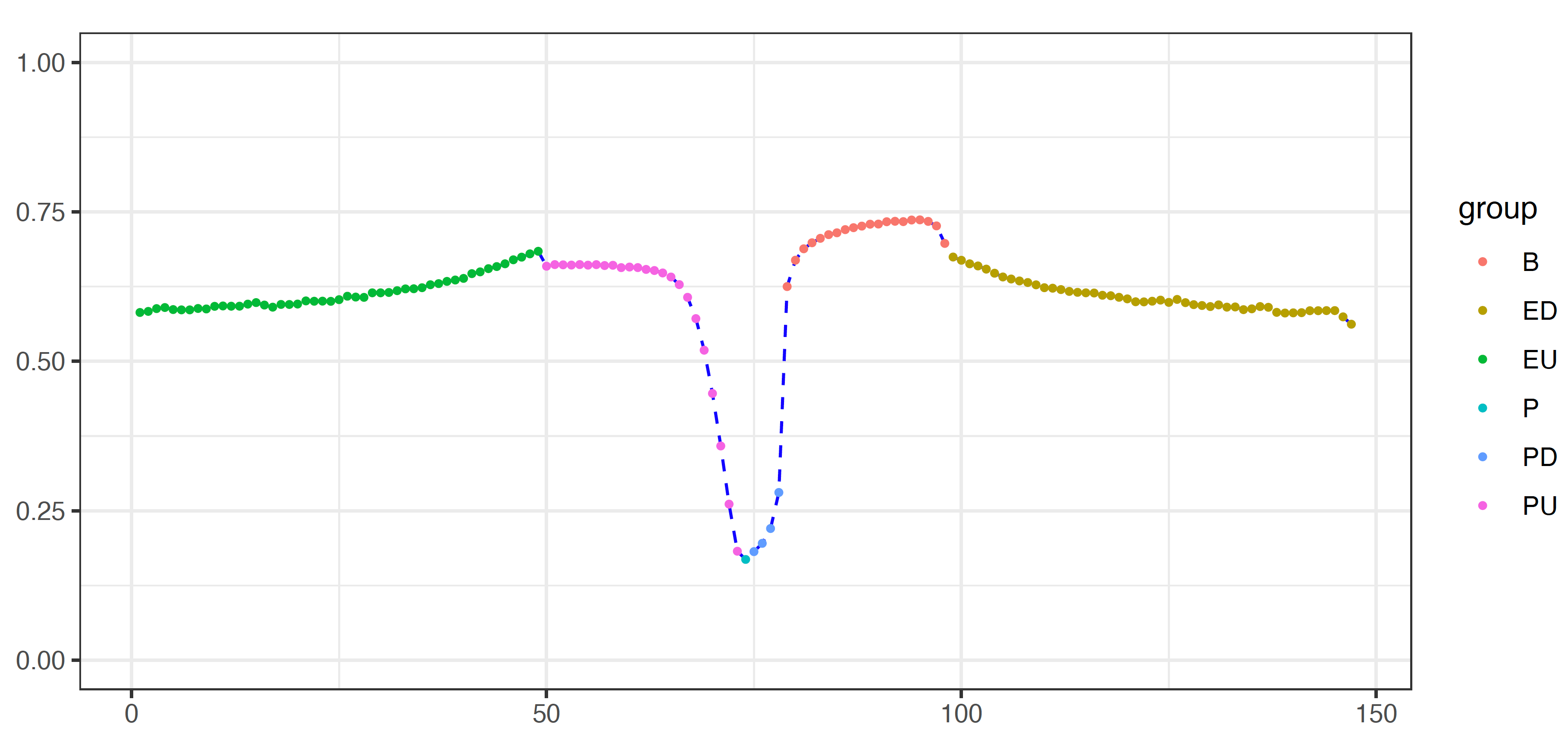
In the 4th column, the unique ID for each region was designed to contain ENSEMBL gene ID, gene symbol, feature and window number for each feature. There are 4 types of features, including distal upstream regions (EU), promoters (P, PU, PD), gene bodies (B) and distal downstream regions (ED).
This can be done using the multiBigwigSummary function in deepTools. Genome-wide signal such as DNA methylation should be stored in a bigwig file.
/sibcb/program/install/deeptools-3.2.1/bin/multiBigwigSummary BED-file \
--BED hg19_windows.bed \
-b esophagus_T_MM.bw \
--outRawCounts esophagus_T_score.tsv \
-o tmp.npz
The resulting file should cover all regions in window file but reordered.
#'chr' 'start' 'end' 'esophagus_T_MM.bw'
chr1 14090 34090 0.5711935773835404
chr1 34090 54090 0.726074748171194
chr1 64190 64390 nan
chr1 64390 64590 0.375
chr1 64590 64790 0.4534412920475006
chr1 64790 64990 0.35458168387413025
chr1 64990 65190 0.9166666865348816
chr1 65190 65390 nan
chr1 65390 65590 nan
Rscript getProfile.R -h
Options:
--windowFile=WINDOWFILE
Window file returned by makeWindows.
--signalFile=SIGNALFILE
Signal file generated by deepTools on regions defined in window_file.
--BED=BED
BED file of interest.
--method=METHOD
Summary method [mean, median].
--outTag=OUTTAG
Prefix for output files.
-h, --help
Show this help message and exit
The signal file was generated in the previous step.
Rscript getProfile.R --windowFile hg19_windows.bed \
--signalFile esophagus_T_score.tsv \
--outTag esophagus_T
Rscript plotProfile.R -h
Options:
--profile_file=PROFILE_FILE
One profile file to be plotted.
--sample_sheet=SAMPLE_SHEET
A samplesheet to describe multiple profile files.
--outTag=OUTTAG
Prefix for output files.
-h, --help
Show this help message and exi
To plot one profile:
Rscript plotProfile.R --profile_file esophagus_T_profile.tsv --outTag esophagus_T
It was known that gene expression are negatively correlated with DNA methylation in promoter regions. However, the relationship is less explored in other features of the genome nor in the presence of other regulatory regions such as open chromatin regions.
singleSampleRA.R -h
Options:
--windowFile=WINDOWFILE
Window file returned by makeWindows.
--signalFile=SIGNALFILE
Signal file generated by deepTools on regions defined in window_file.
--BED=BED
BED file of interest.
--expression_file=EXPRESSION_FILE
Gene expression profile [1st: gene_id/gene_name; 2nd: log transformed value].
--min_N=MIN_N
Minimal number of genes to calculate summary scores [20].
--min_methylation=MIN_METHYLATION
Minimal methylation for further analysis [0].
--max_methylation=MAX_METHYLATION
Maximal methylation for further analysis [1].
--method=METHOD
Summary method [Pearson, Spearman, median, mean].
--outTag=OUTTAG
Prefix for output files.
-h, --help
Show this help message and exit
The method parameter controls how summary is performed. By setting this parameter as Pearson or Spearman, correlation will be calculated between DNA methylation and gene expression for each window. Alternatively, setting this parameter to median or mean will ignore DNA methylation and reported median or mean DNA methylation in each window.
Rscript singleSampleRA.R \
--windowFile hg19_windows.bed \
--signalFile esophagus_T_score.tsv \
--BED esophagus_T_MHB.txt \
--min_methylation 0 \
--max_methylation 1 \
--method Pearson \
--expression_file GSM4505886_T1.txt \
--outTag esophagus_T_Pearson
The results look like:
Num_All Num_Feature Score_All Score_Feature
EU_2 8615 892 0.273584436154975 0.183173404046737
EU_1 10960 1253 0.313645114677926 0.255364543245263
PU_23 14123 84 0.199370638649568 0.0712089652212049
PU_22 14109 86 0.195235920725899 0.0713547901031287
PU_21 14116 88 0.2106527569829 -0.106417868517083
PU_20 14181 72 0.204725214923501 -0.0751451609553298
PU_16 14130 85 0.203080136300442 0.0249686692827358
PU_11 14334 86 0.181644123785359 0.282236463340651
PU_10 14520 108 0.173973677565651 0.221791973016975