The official implementation of the paper "Rolling-Unet: Revitalizing MLP’s Ability to Efficiently Extract Long-Distance Dependencies for Medical Image Segmentation" published in AAAI-2024.
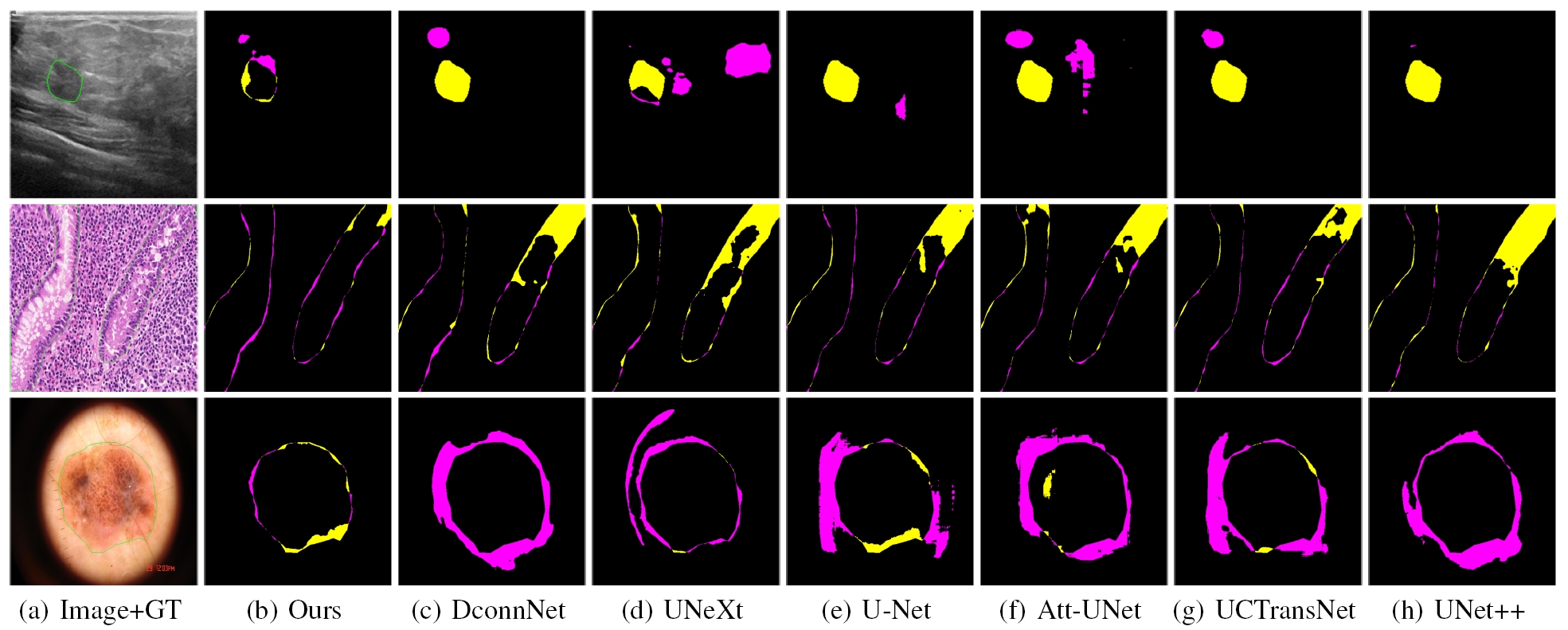
Medical image segmentation methods based on deep learning network are mainly divided into CNN and Transformer. However, CNN struggles to capture long-distance dependencies, while Transformer suffers from high computational complexity and poor local feature learning. To efficiently extract and fuse local features and long-range dependencies, this paper proposes Rolling-Unet, which is a CNN model combined with MLP. Specifically, we propose the core R-MLP module, which is responsible for learning the long-distance dependency in a single direction of the whole image. By controlling and combining R-MLP modules in different directions, OR-MLP and DOR-MLP modules are formed to capture long-distance dependencies in multiple directions. Further, Lo2 block is proposed to encode both local context information and long-distance dependencies without excessive computational burden. Lo2 block has the same parameter size and computational complexity as a 3×3 convolution. The experimental results on four public datasets show that Rolling-Unet achieves superior performance compared to the state-of-the-art methods.
-
The experimental environment of this paper includes: Ubuntu 20.04 LTS system, an i9-12900K CPU, an NVIDIA RTX A6000 GPU, Python version 3.8, Pytorch version 1.13, and CUDA version 11.6.
-
Clone this repository:
git clone https://github.com/Jiaoyang45/Rolling-Unet
cd Rolling-Unet-
In the paper, for the BUSI, GlaS, and CHASEDB1 datasets, we performed three random splits using the "train_test_split" function, with corresponding random seeds of 41, 1029, and 3407, respectively. ISIC 2018 dataset only performed random split once, with a corresponding seed of 41. To ensure the reproducibility of the experiment, we strictly set the random seeds for functions such as Torch, Python, and Numpy to 1029 using the "seed_torch" function.
-
For the CHASEDB1 dataset, The "test_size" of "train_test_split" is not 0.2, but 0.27, which makes the training set exactly 20 images, which is a multiple of batch_size, in order to fully utilize the data for learning. The remaining 8 images are fully utilized for evaluation.
- Make sure to put the files as the following structure. For binary segmentation, just use folder 0.
inputs
└── <dataset name>
├── images
| ├── 001.png
│ ├── 002.png
│ ├── 003.png
│ ├── ...
|
└── masks
└── 0
├── 001.png
├── 002.png
├── 003.png
├── ...
- Train the model.
python train.py
- Evaluate.
python val.py
- This code includes visualization of the training process through tensorboardX, and calculation multiple indicators through MedPy.
This code repository is implemented based on UNeXt.
- UNet, UNet++, Attention-UNet - link
- Medical-Transformer - link
- UCTransNet - link
- UNeXt - link
- DconnNet - link
If this code is helpful for your study, please cite:
- Liu, Y., Zhu, H., Liu, M., Yu, H., Chen, Z., & Gao, J. (2024). Rolling-Unet: Revitalizing MLP’s Ability to Efficiently Extract Long-Distance Dependencies for Medical Image Segmentation. Proceedings of the AAAI Conference on Artificial Intelligence, 38(4), 3819-3827. https://doi.org/10.1609/aaai.v38i4.28173