Knowledge Graph Reasoning for Explainable Prediction of Synthetic Lethality
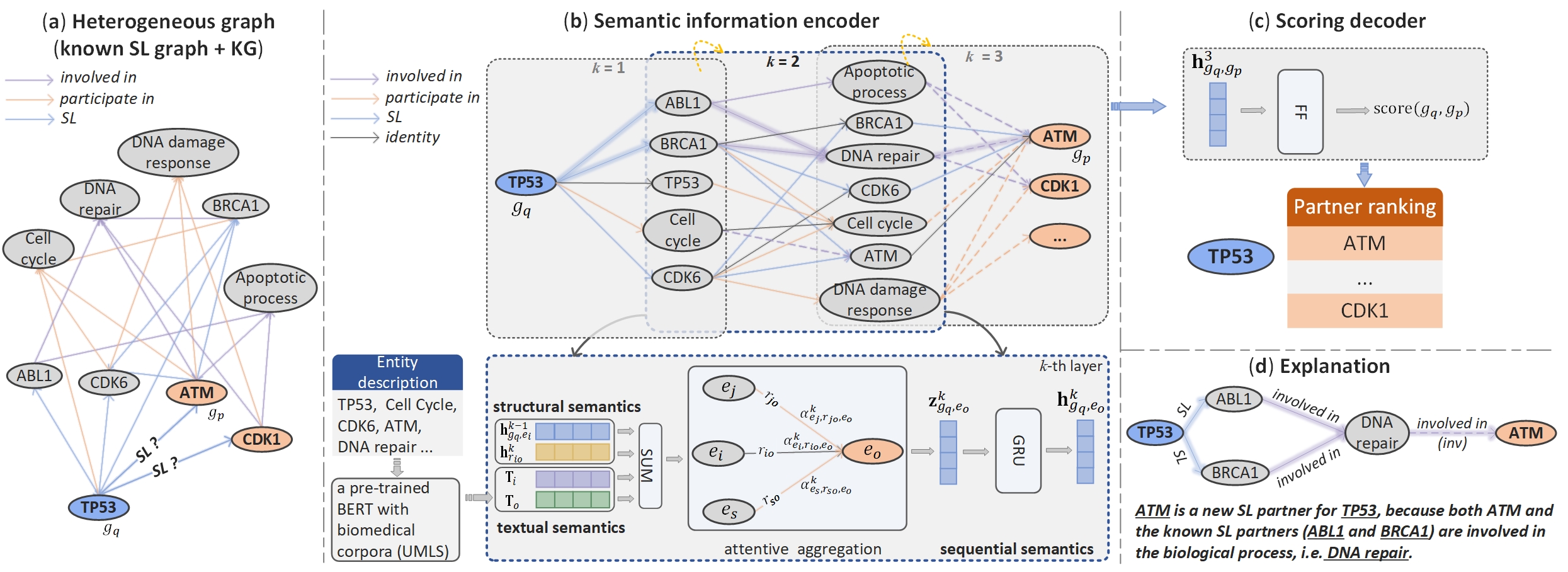
We have provided the processed dataset. The SL gene pairs we used are derived from SynLethDB-v2.0. To construct a knowledge graph containing more relation types, we first use the knowledge about genes and their functions SynLethKG, i.e. five types of entities include Gene, Pathway and three types of GO terms (biological processes (BP), molecular functions (MF) and cellular components (CC)). Four types of relations are included to describe if a gene is involved in a pathway or annotated with a GO term, namely, Participate_in_PW, Participate_in_BP, Participate_in_MF, and Participate_in_CC. We then expand the relation types of gene-GO and GO-GO, using the data from [OntoProtein] (https://github.com/zjunlp/OntoProtein). See kg.txt in data folder for the complete KG.
To utilize the pre-trained textual embedding, one can take all_entities.txt in data folder as the input and use the pre-trained BERT model named CODER to get textual embeddings for each entity.
Extract textual embeddings
python extract_pretrain_emb.py
For transductive reasoning
cd transductive
python -W ignore train.py --suffix trans_reason
For inductive reasoning
cd inductive
python -W ignore train.py --suffix ind_reason
python36
torch 1.11.0
torch scatter 2.0.9
transformers 4.23.1
Partial code is adapted from Zhang and Yao 2022 with the repository https://github.com/AutoML-Research/RED-GNN.