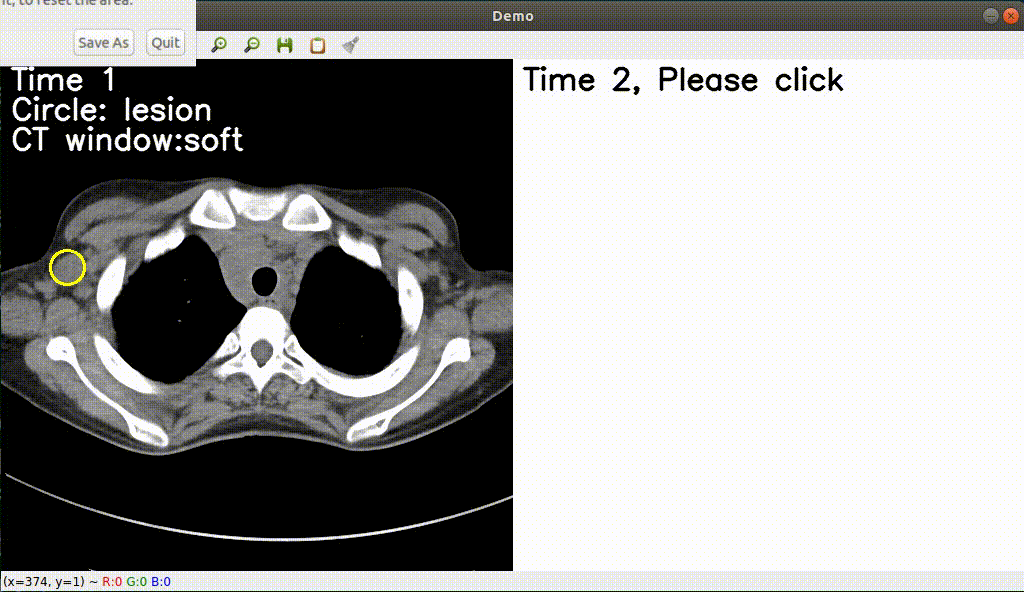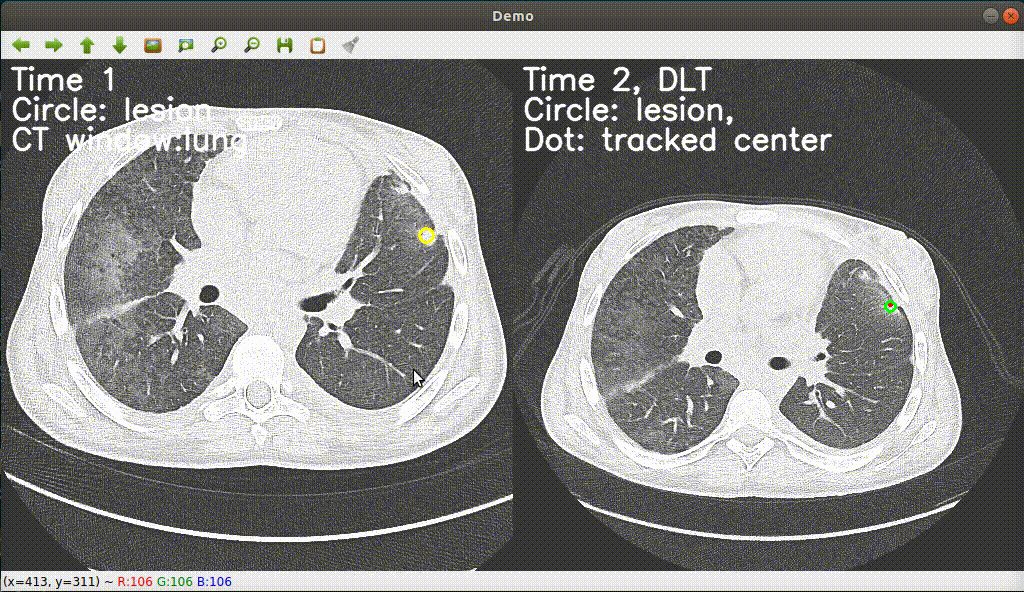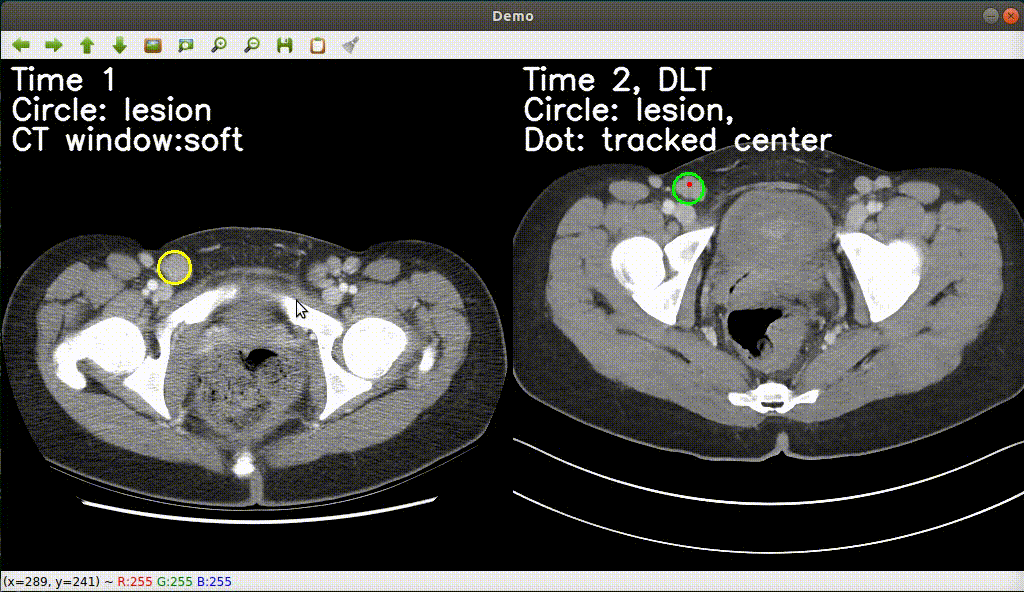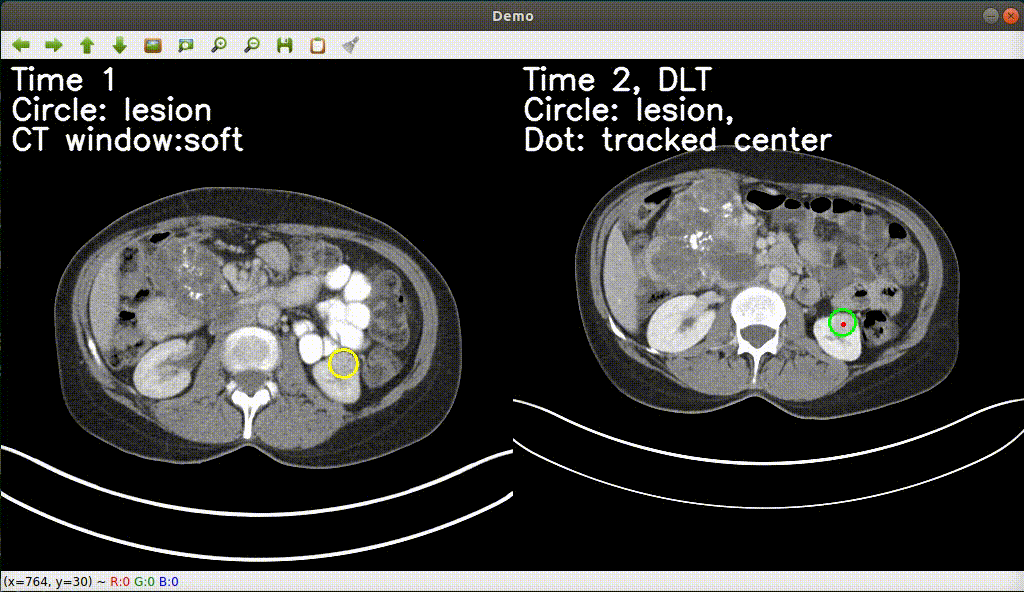
Monitoring treatment response in longitudinal studies plays an important role in clinical practice. Accurately identifying lesions across serial imaging follow-up is the core to the monitoring procedure. Typically this incorporates both image and anatomical considerations. However, matching lesions manually is labor-intensive and time-consuming. In this project, we release our lesion tracking benchmark -- deep longitudinal study (DLS) dataset, consisting of 3891 lesion pairs from the public DeepLesion database. With DLS, we develop our deep lesion tracker (DLT) to efficiently and accurately track lesions in longitudinal studies.
Deep Lesion Tracker: Monitoring Lesions in 4D Longitudinal Imaging Studies
Jinzheng Cai, Youbao Tang, Ke Yan, Adam P. Harrison, Jing Xiao, Gijin Lin, Le Lu
will show in CVPR2021
Contact: caijinzhengcn@gmail.com from PAII Inc. Any questions or discussions are welcomed!
-
Annotation: you can find annotation files in the data folder. The structure of annotation is defined as:
import json train = json.load(open('./data/train.json', 'r')) # List of the below lesion_pair_annotation lesion_pair_annotation = { 'source': , # name of the source image, xxx.nii.gz 'target': , # name of the target image, xxx.nii.gz 'source box': , # 3D bounding box of the source lesion, [xmin, ymin, zmin, width, height, depth] 'target box': , # 3D bounding box of the target lesion, [xmin, ymin, zmin, width, height, depth] 'source center': , # center of the source lesion, [x, y, z] 'target center': , # center of the target lesion, [x, y, z] 'predict target center': , # affine initialized center of the target lesion, [x, y, z] 'predict target box': , # affine initialized box of the target lesion, [xmin, ymin, zmin, width, height, depth] 'source spacing': , # CT spacing of the source image 'target spacing': , # CT spacing of the target image 'source recist slice': , # name of the RECIST slice, xxx/xxx.png 'source recist coordinate': , # end points of the source RECIST mark, [p1x, p1y, p2x, p2y, p3x, p3y, p4x, p4y] 'source recist diameter': , # diameters of the source lesion in mm, [long axis, short axis] 'source recist box': , # 2D bounding box of the source lesion on the RECIST slice, [xmin, ymin, width, height] 'source recist center': , # center of the source RECIST mark, [x, y, z], same as 'source center' 'target recist slice': , # name of the RECIST slice, xxx/xxx.png 'target recist coordinate': , # end points of the target RECIST mark, [p1x, p1y, p2x, p2y, p3x, p3y, p4x, p4y] 'target recist diameter': , # diameters of the target lesion in mm, [long axis, short axis] 'target recist box': , # 2D bounding box of the target lesion on the RECIST slice, [xmin, ymin, width, height] 'target recist center': , # center of the target RECIST mark, [x, y, z], same as 'target center' }
-
Data: in order to use our annotations, you need to convert DeepLesion from png to nifti:
Please download DL_save_nifti.py from the official website of DeepLesion. Then run,python DL_save_nifti.py
It generates CT subvolumes named in the format of PatientID_StudyID_ScanID_StartingSliceID_EndingSliceID.nii.gz, for example "001344_01_01_012-024.nii.gz".
-
Evaluation: find example evaluation code:
python evaluation.py
DLT is released under the CC-BY-SA-4.0 License (refer to the LICENSE file for details).
If you find this project useful for your research, please use the following BibTeX entry.
@inproceedings{cai2020deep,
title={Deep Lesion Tracker: Monitoring Lesions in 4D Longitudinal Imaging Studies},
author={Jinzheng Cai and Youbao Tang and Ke Yan and Adam P. Harrison and Jing Xiao and Gigin Lin and Le Lu},
booktitle={CVPR},
year={2021}
}