Tools to compare structural variants to those in public databases!
Publication: https://f1000research.com/articles/5-673/v1
#Introduction:
NCBI’s dbVar database of large structural variation (https://www.ncbi.nlm.nih.gov/dbvar) contains over 3 million human submitted structural variants, including variants with clinical assertions from ClinVar. These data were submitted from over 120 human studies, including 1000 Genomes, and include copy number variants (CNVs), insertions, deletions, duplications, inversions, and translocations. This wealth of human SV data represents an important resource that can be used to help identify normal and causal variants among the millions of genomes that will be sequenced during the next few years in the fields of personal genomics, medical genetics, and variant discovery.
Despite the value such a large, centralized collection of human SV represents, it is challenging to combine the data and package them in a form which users can usefully and easily incorporate into analysis pipelines. Confounding factors include varying levels of variant qa and validation, dbVar’s study-centric organization, unavoidable variant breakpoint ambiguity, and remapping hurdles in difficult regions of the genome. To address this Big Data problem, participants in a dbVar NCBI Hackathon (January 4-6, 2016) collaborated to produce structural variant tools and dbVar datasets that aim to:
- facilitate the comparison of dbVar SV data with other genome annotations, such as disease phenotype and population frequencies
- improve data exchange, mining, computation, and reporting
- improve searching and matching of genomic coordinates across studies and assemblies
- simplify the display of dbVar SV in genome browsers, as aggregated histograms or density tracks organized by variant type, study, or assembly
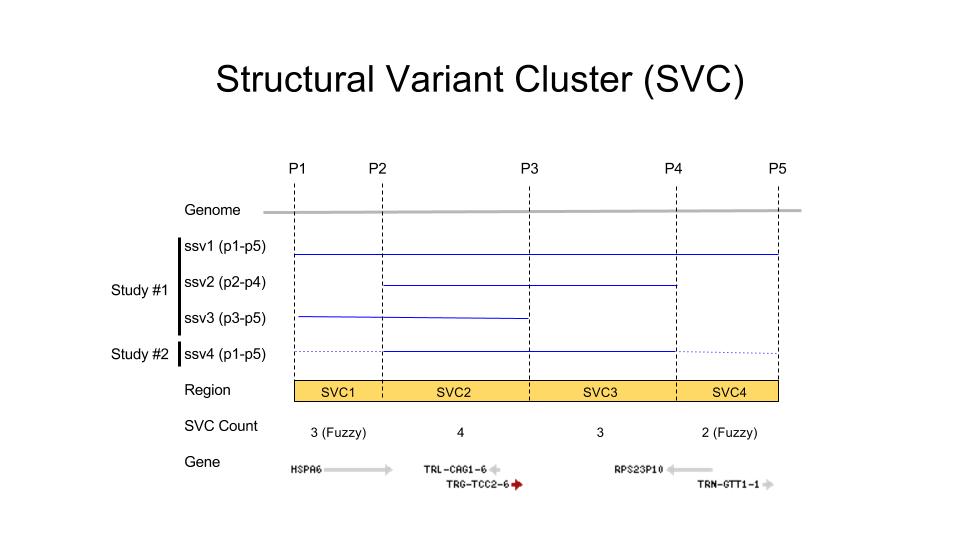
The Hackathon team generated a nonredundant set of genomic regions called Structural Variant Clusters (SVC). These features are defined by regions of concordance amongst submitted human SSVs placed on assembly GRCh38 (see Figure 1). One can easily compare these SVC data against other annotated genomic regions such as segmental duplications, clinically significant genes, dosage sensitive and/or essential regions, and problematic genomic regions that may yield suspected false variants. We also developed open-source utilities to: 1) search and filter SVC data in GVF format, 2) compute summary statistics and export data for genomic viewers, and 3) annotate SVCs using external data sources.
Installation and Test instructions:
Data:
Human dbVar study input tab files based on GRCh38 ftp://ftp.ncbi.nlm.nih.gov/pub/dbVar/data/Homo_sapiens/by_study/
Complete listing of all organisms available in dbVar and recent updates (http://www.ncbi.nlm.nih.gov/dbvar/content/ftp_manifest/) for input
Precomputed SVC by study (January 2016) ftp://ftp.ncbi.nlm.nih.gov/pub/dbVar/sandbox/hackathon/January_2016/
More Info
- Presentation: https://docs.google.com/presentation/d/1n6WslYsnFPs74du63Y8hXgTj8PKH5b5V_QC9GqDH0Oo/edit?usp=sharing
- Structural_Variant_Comparison wiki: https://github.com/NCBI-Hackathons/Structural_Variant_Comparison/wiki
- Last updated: April 4, 2016