Code accompanying Explaining the Model and Feature Dependencies by Decomposition of the Shapley Value.
SHAP values have become increasingly popular in recent years. However, it is (was) unclear how to apply them to datasets
with dependent features. In our paper we provide a summary of the different options that arise and their
motivations. Essentially, two popular SHAP implementations exist with followers on both sides: one that explains the
model (Interventional SHAP [1], the regular shap implementation) and one that
explain the model combined with the feature dependencies (Conditional SHAP [2]). We will illustrate that both
options are
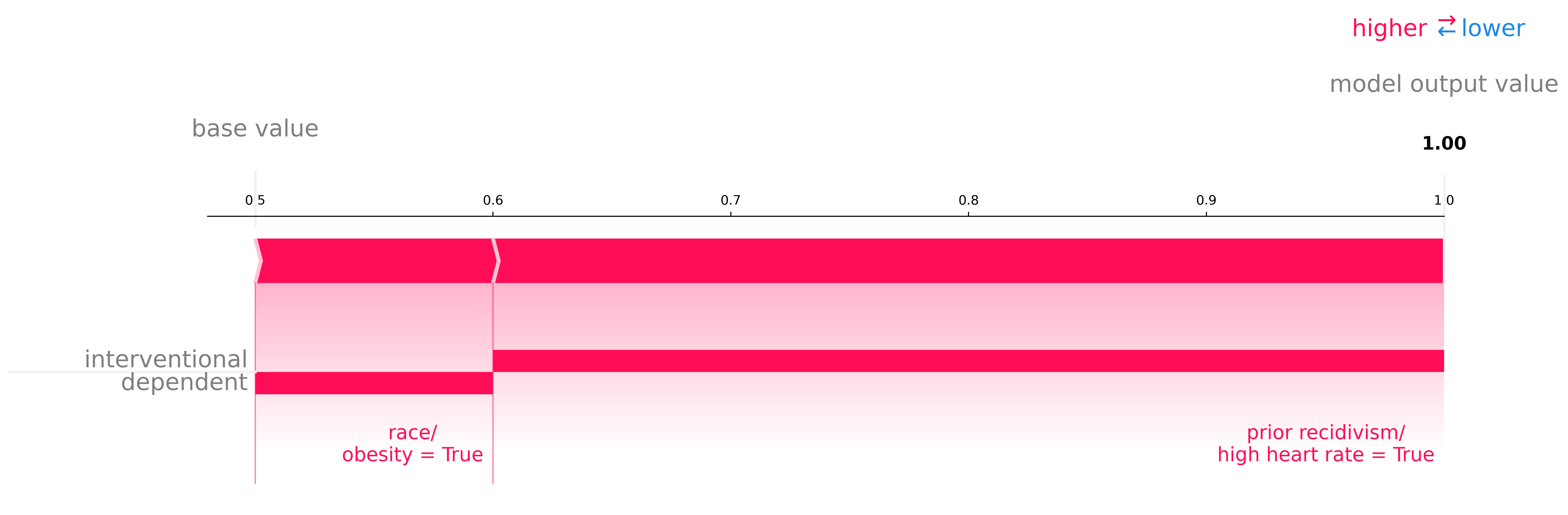
valuable for different inferences with a simple example. Consider two binary dependent features
| Interventional SHAP | Conditional SHAP | Decomposition SHAP | ||||
|---|---|---|---|---|---|---|
| court | recidivist | race | future recidivism | ✔️ | ✔️ | |
| hospital | high heart rate | obesity | heart attack | ✔️ | ✔️ |
Given a certain sample, with SHAP we explain its output by determining what effect every feature value has towards
the output. In the first setting
Preferably, a good method should provide both inferences. Therefore, we propose to split up a SHAP value for a feature into a model corresponding (interventional) attribution and an attribution via other dependent features, allowing the end-user (e.g. convict or patient) to easily grasp the explanation of both the model and feature dependencies.
These attributions can be visualized through our
modified force plots. For the simple toy example above, we explain
We see that the convict's race has no interventional effect. Whether the patient is obese does have an output effect via its dependent feature high heart rate which is clearly shown in the explanation.
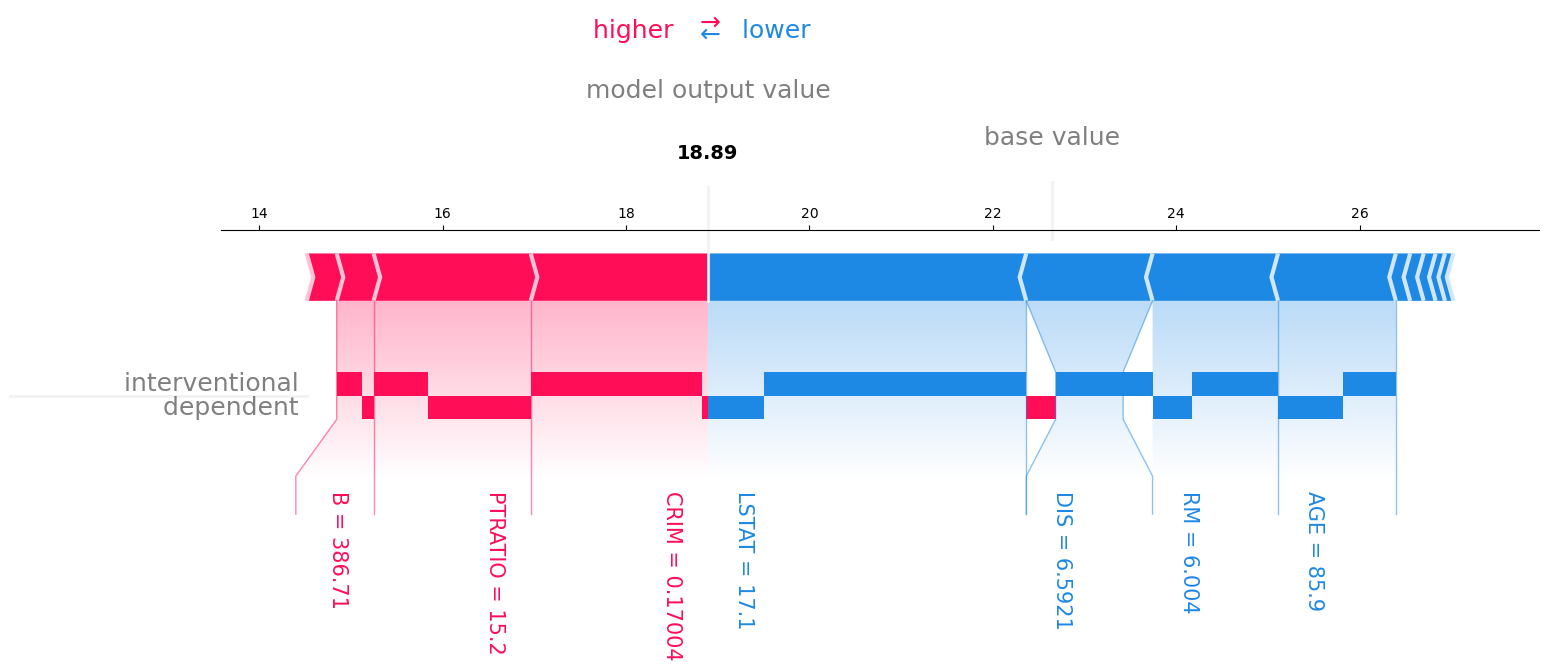
Furthermore, we can apply our method to the standard Boston Housing dataset and explain the prediction of a random forest model.
We see that the crime rate per capita (
- clone the Github respository
- install Conda environment (requires Conda):
conda env create -f environment.yml
Let's illustrate the use of our toolbox with an example. We will use the Breast Cancer Wisconsin dataset
and a XGBoost model to predict the cancer type. We will then explain the prediction of a test sample using
Decomposition SHAP. The full code is available in example.py.
First load the data and train the model:
import numpy as np
import xgboost as xgb
from sklearn.datasets import load_breast_cancer
from sklearn.model_selection import train_test_split
from decomposition_shap.distributions import MultiGaussian
from decomposition_shap.force_dependent import force_dependent_plot
from decomposition_shap.kernel_dependent import DependentKernelExplainer
np.random.seed(0)
# Load Breast Cancer Wisconsin dataset
x, y = load_breast_cancer(return_X_y=True)
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
# Fit a model
model = xgb.XGBClassifier(n_estimators=10)
model.fit(x_train, y_train)Since we are computing data dependencies, an assumed data distribution is required. In this case we will use a multivariate Gaussian distribution. We can now explain the prediction of a test sample:
# Fit feature distribution (assumed Multivariate Gaussian)
dist = MultiGaussian(x_train)
# Note: custom distributions can also be defined see decomposition_shap/distribution.py
# Initialize explainer
explainer = DependentKernelExplainer(lambda d: model.predict(d, output_margin=True), x_train, dist.sample)
# Note: as with regular SHAP, usually it makes more sense to explain the logodds 'model.predict(d, output_margin=True)'
# instead of the class probabilities 'model.predict(d)'
# Explain the third test sample
sample_index = 3
shaps = explainer.shap_values(x_test[sample_index]) # returns array of conditional SHAP values + interventional effectsThe SHAP values can be visualized using the force_dependent_plot function:
data = load_breast_cancer()
# With link='logit' the logodds are shown under the axis, and the probability is shown above the axis
fig = force_dependent_plot(np.mean(model.predict(x_train)), shaps, features=x_test[sample_index],
feature_names=data['feature_names'], link='logit', text_rotation=-90) # returns plt.gcf()
fig.savefig("example.png", bbox_inches="tight")\decomposition_shap\kernel_dependent.pyholds functions that can compute interventional and dependent effects- class
DependentKernelExplainerexplains output of any function for any situation by decomposing the Shapley value into an interventional and dependent effect. See code documentation for information on constructor arguments. - function
shap_values(...)estimates the SHAP values and interventional effects for a set of samples. See code documentation for information on arguments.
- class
\decomposition_shap\force_dependent.pyholds implementation of force plot visualizations with interventional and dependent effects- function
force_dependent_plot(...)visualizes the given SHAP values and interventional effects in a matplotlib force plot. See code documentation for information on arguments.
- function
\decompostion_shap\distributions.pyholds different implementations of assumed data distributions\experimentshas all code to do all experiments of the paper and generate all resultsexample.pyhas a simple example to illustrate the use of Decomposition SHAPteststests of the implemented code
If you use Decomposition SHAP in your research we would appreciate citing Explaining the Model and Feature Dependencies by Decomposition of the Shapley Value
[1] Janzing, D., Minorics, L., & Blöbaum, P. (2020, June). Feature relevance quantification in explainable AI: A causal problem. In International Conference on artificial intelligence and statistics (pp. 2907-2916). PMLR.
[2] Aas, K., Jullum, M., & Løland, A. (2021). Explaining individual predictions when features are dependent: More accurate approximations to Shapley values. Artificial Intelligence, 298, 103502.