K-means clustering is a type of unsupervised learning, which is used when you have unlabeled data (i.e., data without defined categories or groups). The goal of this algorithm is to find groups in the data, with the number of groups represented by the variable K. The algorithm works iteratively to assign each data point to one of K groups based on the features that are provided. Data points are clustered based on feature similarity. The results of the K-means clustering algorithm are:
The centroids of the K clusters, which can be used to label new data
Labels for the training data (each data point is assigned to a single cluster)
Rather than defining groups before looking at the data, clustering allows you to find and analyze the groups that have formed organically. The "Choosing K" section below describes how the number of groups can be determined.
Each centroid of a cluster is a collection of feature values which define the resulting groups. Examining the centroid feature weights can be used to qualitatively interpret what kind of group each cluster represents.
The Κ-means clustering algorithm uses iterative refinement to produce a final result. The algorithm inputs are the number of clusters Κ and the data set. The data set is a collection of features for each data point. The algorithms starts with initial estimates for the Κ centroids, which can either be randomly generated or randomly selected from the data set. The algorithm then iterates between two steps:
- Data assigment step:
Each centroid defines one of the clusters. In this step, each data point is assigned to its nearest centroid, based on the squared Euclidean distance. More formally, if ci is the collection of centroids in set C, then each data point x is assigned to a cluster based on
where dist( · ) is the standard (L2) Euclidean distance. Let the set of data point assignments for each ith cluster centroid be Si.
- Centroid update step:
In this step, the centroids are recomputed. This is done by taking the mean of all data points assigned to that centroid's cluster.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cluster import KMeansdf = pd.read_csv('vinos.csv')df.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 178 entries, 0 to 177
Data columns (total 14 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Vino 178 non-null int64
1 Alcohol 178 non-null float64
2 Malic 178 non-null float64
3 Ash 178 non-null float64
4 Alcalinity 178 non-null float64
5 Magnesium 178 non-null int64
6 Phenols 178 non-null float64
7 Flavanoids 178 non-null float64
8 Nonflavanoids 178 non-null float64
9 Proanthocyanins 178 non-null float64
10 Color 178 non-null float64
11 Hue 178 non-null float64
12 Dilution 178 non-null float64
13 Proline 178 non-null int64
dtypes: float64(11), int64(3)
memory usage: 19.6 KB
df.head()
df.drop(['Vino'], axis=1, inplace=True)df.describe().dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| Alcohol | Malic | Ash | Alcalinity | Magnesium | Phenols | Flavanoids | Nonflavanoids | Proanthocyanins | Color | Hue | Dilution | Proline | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 |
| mean | 13.000618 | 2.336348 | 2.366517 | 19.494944 | 99.741573 | 2.295112 | 2.029270 | 0.361854 | 1.590899 | 5.058090 | 0.957449 | 2.611685 | 746.893258 |
| std | 0.811827 | 1.117146 | 0.274344 | 3.339564 | 14.282484 | 0.625851 | 0.998859 | 0.124453 | 0.572359 | 2.318286 | 0.228572 | 0.709990 | 314.907474 |
| min | 11.030000 | 0.740000 | 1.360000 | 10.600000 | 70.000000 | 0.980000 | 0.340000 | 0.130000 | 0.410000 | 1.280000 | 0.480000 | 1.270000 | 278.000000 |
| 25% | 12.362500 | 1.602500 | 2.210000 | 17.200000 | 88.000000 | 1.742500 | 1.205000 | 0.270000 | 1.250000 | 3.220000 | 0.782500 | 1.937500 | 500.500000 |
| 50% | 13.050000 | 1.865000 | 2.360000 | 19.500000 | 98.000000 | 2.355000 | 2.135000 | 0.340000 | 1.555000 | 4.690000 | 0.965000 | 2.780000 | 673.500000 |
| 75% | 13.677500 | 3.082500 | 2.557500 | 21.500000 | 107.000000 | 2.800000 | 2.875000 | 0.437500 | 1.950000 | 6.200000 | 1.120000 | 3.170000 | 985.000000 |
| max | 14.830000 | 5.800000 | 3.230000 | 30.000000 | 162.000000 | 3.880000 | 5.080000 | 0.660000 | 3.580000 | 13.000000 | 1.710000 | 4.000000 | 1680.000000 |
df_norm = (df - df.min())/(df.max()-df.min())
df_norm.head().dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| Alcohol | Malic | Ash | Alcalinity | Magnesium | Phenols | Flavanoids | Nonflavanoids | Proanthocyanins | Color | Hue | Dilution | Proline | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.842105 | 0.191700 | 0.572193 | 0.257732 | 0.619565 | 0.627586 | 0.573840 | 0.283019 | 0.593060 | 0.372014 | 0.455285 | 0.970696 | 0.561341 |
| 1 | 0.571053 | 0.205534 | 0.417112 | 0.030928 | 0.326087 | 0.575862 | 0.510549 | 0.245283 | 0.274448 | 0.264505 | 0.463415 | 0.780220 | 0.550642 |
| 2 | 0.560526 | 0.320158 | 0.700535 | 0.412371 | 0.336957 | 0.627586 | 0.611814 | 0.320755 | 0.757098 | 0.375427 | 0.447154 | 0.695971 | 0.646933 |
| 3 | 0.878947 | 0.239130 | 0.609626 | 0.319588 | 0.467391 | 0.989655 | 0.664557 | 0.207547 | 0.558360 | 0.556314 | 0.308943 | 0.798535 | 0.857347 |
| 4 | 0.581579 | 0.365613 | 0.807487 | 0.536082 | 0.521739 | 0.627586 | 0.495781 | 0.490566 | 0.444795 | 0.259386 | 0.455285 | 0.608059 | 0.325963 |
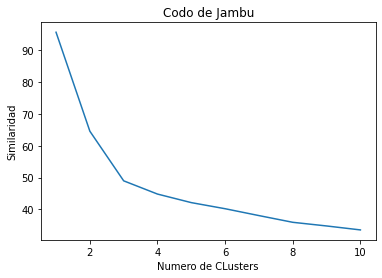
Codo de Jambu: Usar diferentes cluster e ir viendo que tan similares son los individuos dentro de los clusters
similaridad = []
for i in range(1,11):
kmeans = KMeans(n_clusters = i, max_iter = 300)
kmeans.fit(df_norm)
similaridad.append(kmeans.inertia_)plt.plot(range(1,11), k)
plt.title('Codo de Jambu')
plt.xlabel('Numero de CLusters')
plt.ylabel('Similaridad')Text(0, 0.5, 'Similaridad')
clustering = KMeans(n_clusters=3, max_iter=300)
clustering.fit(df_norm)KMeans(n_clusters=3)
df['Cluster'] = clustering.labels_
df.head().dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| Alcohol | Malic | Ash | Alcalinity | Magnesium | Phenols | Flavanoids | Nonflavanoids | Proanthocyanins | Color | Hue | Dilution | Proline | Cluster | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 14.23 | 1.71 | 2.43 | 15.6 | 127 | 2.80 | 3.06 | 0.28 | 2.29 | 5.64 | 1.04 | 3.92 | 1065 | 0 |
| 1 | 13.20 | 1.78 | 2.14 | 11.2 | 100 | 2.65 | 2.76 | 0.26 | 1.28 | 4.38 | 1.05 | 3.40 | 1050 | 0 |
| 2 | 13.16 | 2.36 | 2.67 | 18.6 | 101 | 2.80 | 3.24 | 0.30 | 2.81 | 5.68 | 1.03 | 3.17 | 1185 | 0 |
| 3 | 14.37 | 1.95 | 2.50 | 16.8 | 113 | 3.85 | 3.49 | 0.24 | 2.18 | 7.80 | 0.86 | 3.45 | 1480 | 0 |
| 4 | 13.24 | 2.59 | 2.87 | 21.0 | 118 | 2.80 | 2.69 | 0.39 | 1.82 | 4.32 | 1.04 | 2.93 | 735 | 0 |
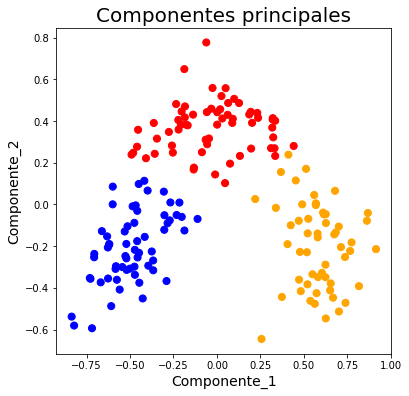
from sklearn.decomposition import PCA
pca = PCA(n_components=2)
pca_vinos = pca.fit_transform(df_norm)
pca_vinos_df = pd.DataFrame(data=pca_vinos, columns=['Componente_1', 'Componente_2'])
pca_nombres_vinos = pd.concat([pca_vinos_df, df[['Cluster']]], axis=1)fig = plt.figure(figsize=(6,6))
ax = fig.add_subplot(1,1,1)
ax.set_xlabel('Componente_1', fontsize=14)
ax.set_ylabel('Componente_2', fontsize=14)
ax.set_title('Componentes principales', fontsize=20)
colores = np.array(['blue','red','orange'])
ax.scatter(x = pca_nombres_vinos.Componente_1, y=pca_nombres_vinos.Componente_2, c=colores[pca_nombres_vinos.Cluster], s=50)<matplotlib.collections.PathCollection at 0x7f81cf4831d0>
df.to_csv('vinos_kmeans.csv')