An R package containing all the functions I have developed to make plotting in R a little easier
Package can be directly installed into R using:
install.packages("devtools")
devtools::install_github("JosephCrispell/basicPlotteR")
library(basicPlotteR)
basicPlotteR is a collection of tools designed for particular tasks. As I develop different tools that help with plotting in R, I'll add them into this general package. These are the current tools available in plotteR:
addTextLabelsfor adding non-overlapping labels onto existing R plotspreadPointsfor adding points onto boxplot(s) that are deterministically spread out to minimise overlapplotMultipleHistogramsfor plotting multiple histograms on the same plotprogressfor running a progress bar within a for loopsetAlphafor changing the alpha value (transparency) of colours specified as strings of characterswatermarkfor adding a transparent text label onto an existing plotplotFASTAa quick way to visualise a nucleotide alignmentaddPointsfor adding non-overlapping points onto existing R plotradarChartfor create a skills radar chart to map skills status and progress
# Create some random points
n <- 50
testLabels <- c("short", "mediummm", "looooonnnnnnngggggg", "0090292002", "9", "A Different label")
coords <- data.frame(X=runif(n), Y=runif(n, min=0, max=100), Name=sample(testLabels, size=n, replace=TRUE),
stringsAsFactors = FALSE)
# Plot them without labels
plot(x=coords$X, y=coords$Y, pch=19, bty="n", xaxt="n", yaxt="n", col="red", xlab="X", ylab="Y")
# With potentially overlapping labels
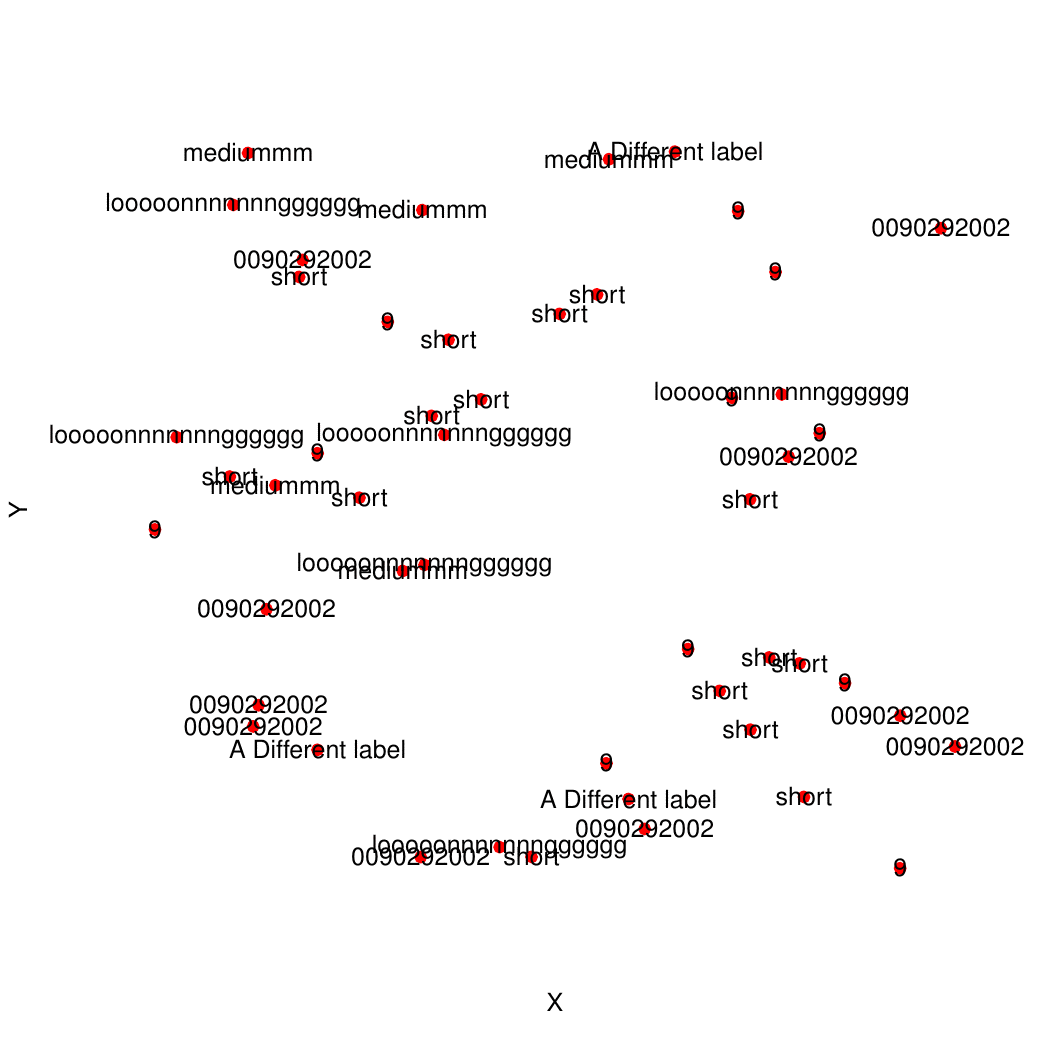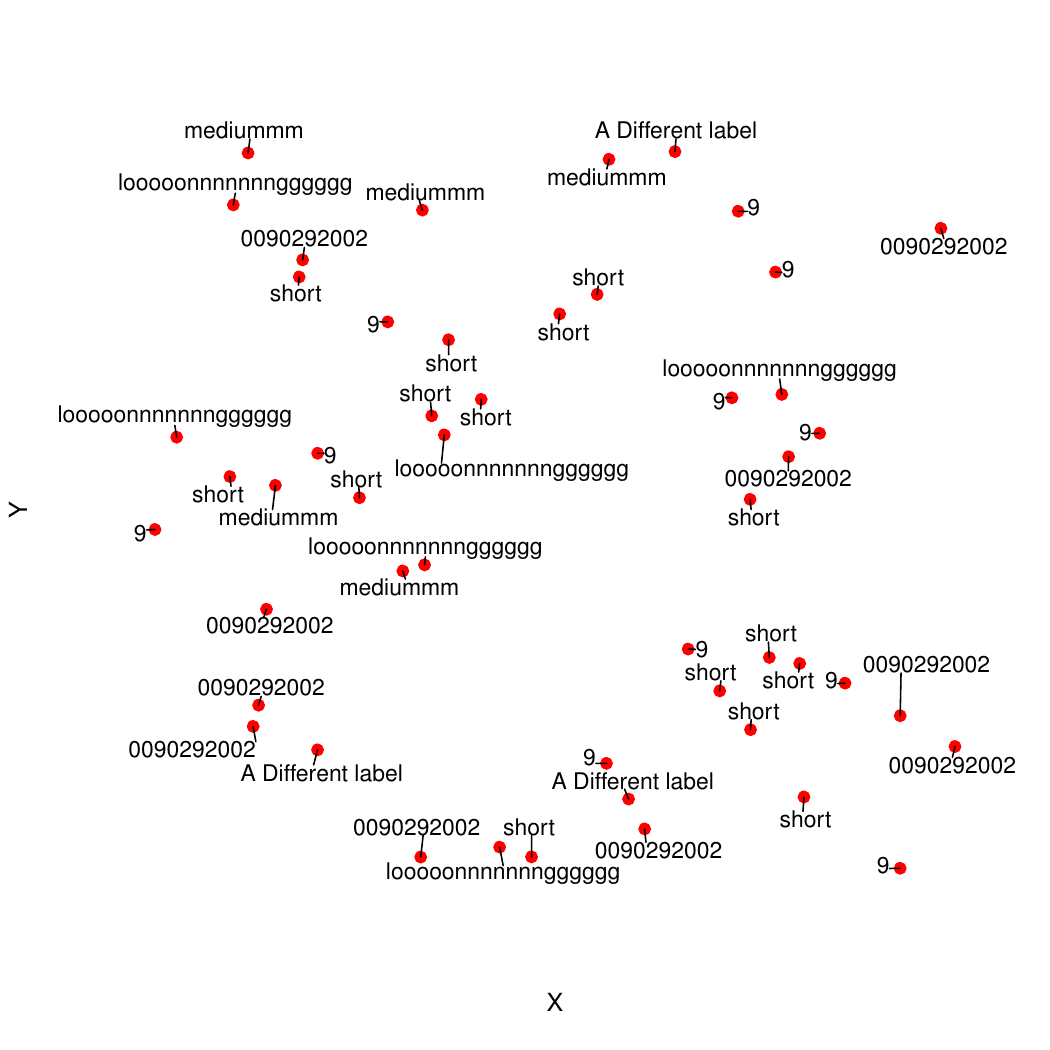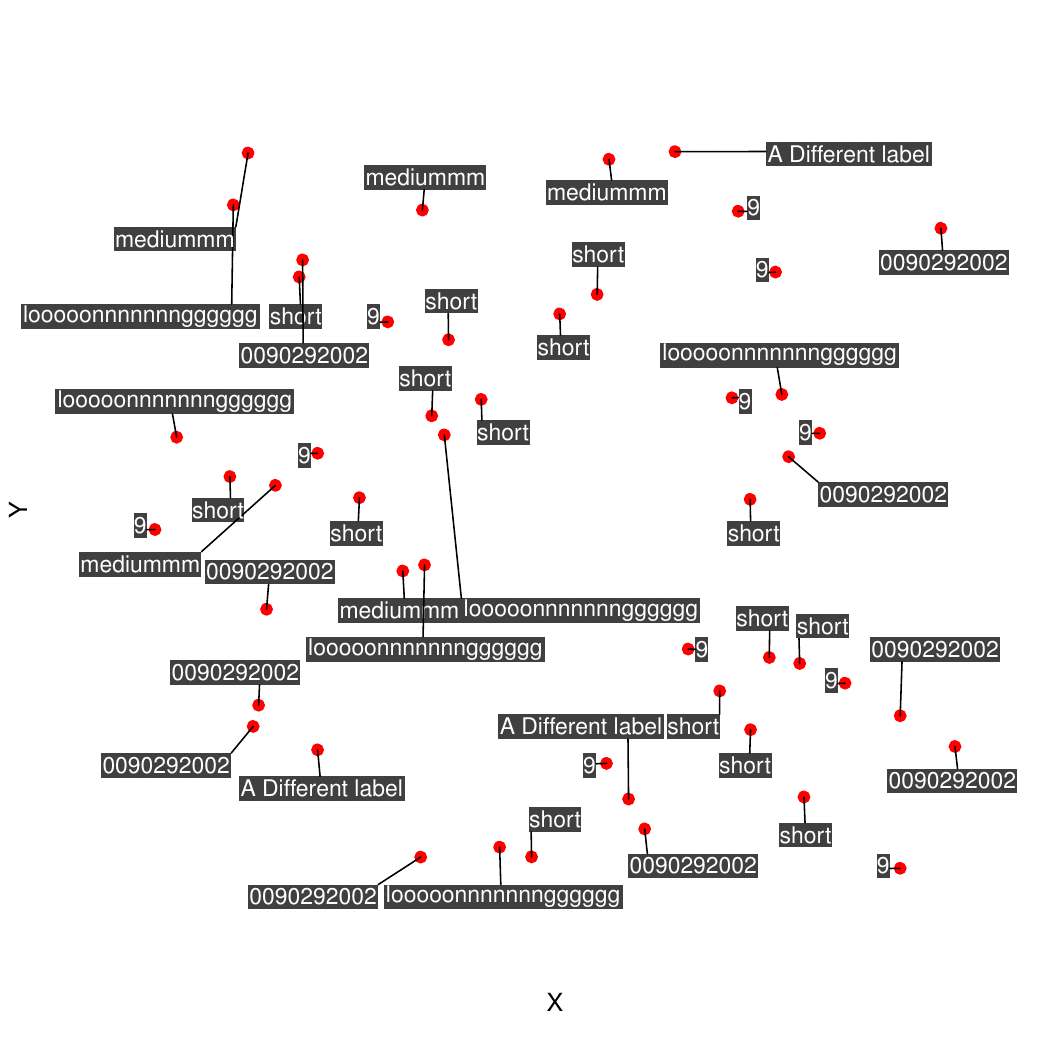
plot(x=coords$X, y=coords$Y, pch=19, bty="n", xaxt="n", yaxt="n", col="red", xlab="X", ylab="Y")
text(coords$X, coords$Y, labels=coords$Name, xpd=TRUE)
# Plot them with non-overlapping labels
plot(x=coords$X, y=coords$Y, pch=19, bty="n", xaxt="n", yaxt="n", col="red", xlab="X", ylab="Y")
addTextLabels(coords$X, coords$Y, coords$Name, cex=1, col.label="black")
# Plot them with non-overlapping labels
plot(x=coords$X, y=coords$Y, pch=19, bty="n", xaxt="n", yaxt="n", col="red", xlab="X", ylab="Y")
addTextLabels(coords$X, coords$Y, coords$Name, cex=1, col.background=rgb(0,0,0, 0.75), col.label="white")
Adding points to single boxplot:
# Set the seed
set.seed(254534)
# Generate some example points - drawn from exponential distribution
values <- rexp(n=50, rate=2)
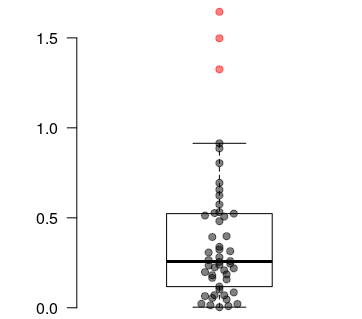
# Plot a boxplot
boxplot(values, xlab="", ylab="", frame=FALSE, las=1, pch=19, outcol=rgb(1,0,0, 0.5),
horizontal=FALSE)
# Plot the points spread along the X axis
spreadPoints(values, position=1)
Adding points to multiple boxplots:
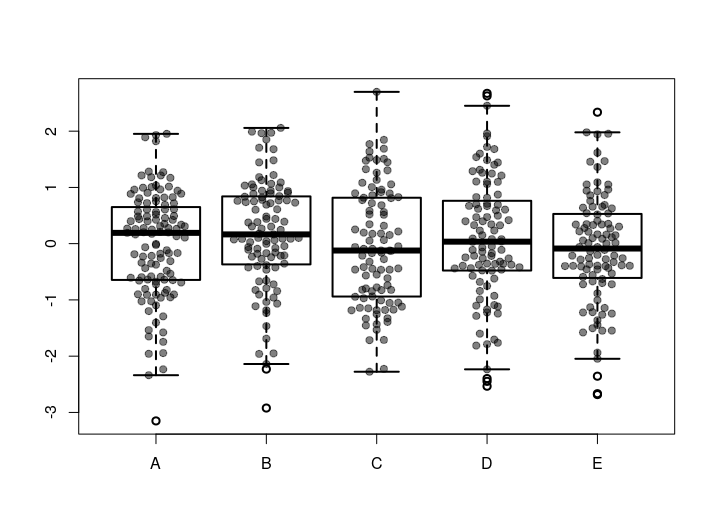
# Set the seed
set.seed(254534)
# Generate some example points - drawn from normal distribution and randomly assign them to categories
randomSamples <- data.frame(Values = rnorm(500), Category = sample(c('A', 'B', 'C', 'D', 'E'), size=500, replace=TRUE))
# Plot a boxplot of the samples from the normal distribution versus there categories - multiple boxplots
boxplot(Values ~ Category, data = randomSamples, lwd = 2)
# Plot the points for each category spread along the X axis
spreadPointsMultiple(data=randomSamples, responseColumn="Values", categoriesColumn="Category")
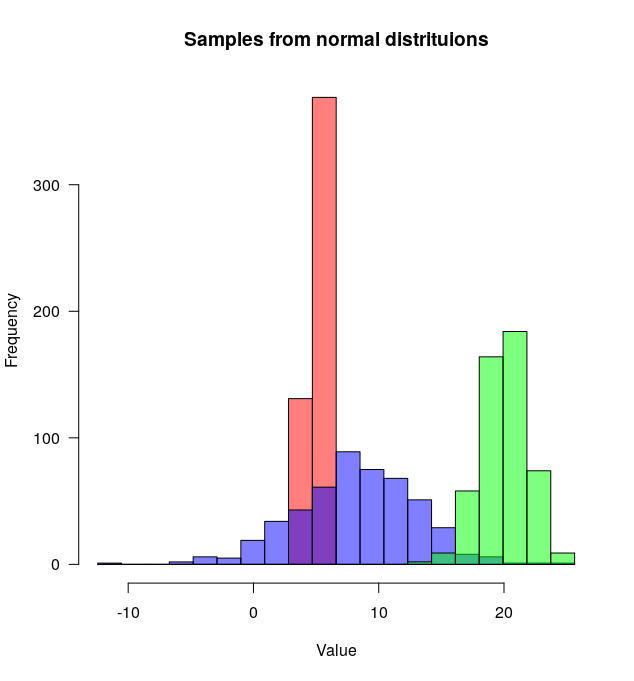
# Set the seed
set.seed(254534)
# Create random samples from a normal distribution
distributions <- list(rnorm(500, mean=5, sd=0.5),
rnorm(500, mean=8, sd=5),
rnorm(500, mean=20, sd=2))
# Plot overlapping histograms
plotMultipleHistograms(distributions, nBins=20, colours=c(rgb(1,0,0, 0.5), rgb(0,0,1, 0.5), rgb(0,1,0, 0.5)),
las=1, main="Samples from normal distribution", xlab="Value")
# Set the number of iterations of the for loop
n <- 1000
for(i in 1:n){
# Sleep for a tenth of a second
Sys.sleep(0.01)
# Update the progress bar
progress(i, n)
}
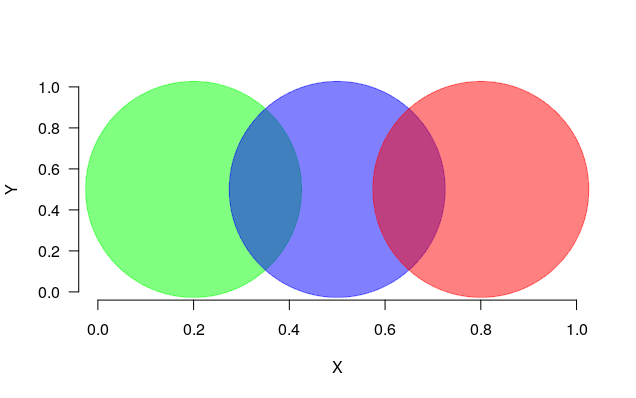
# Define the coordinates of three points
x <- c(0.2, 0.5, 0.8)
y <- c(0.5, 0.5, 0.5)
# Define the colours for the points
colours <- c("green", "blue", "red")
# Plot the points - set the transparency
plot(x, y, bty="n", xlab="X", ylab="Y", las=1, pch=19, cex=30, xlim=c(0,1), ylim=c(0,1),
col=setAlpha(colours, alpha=0.5))
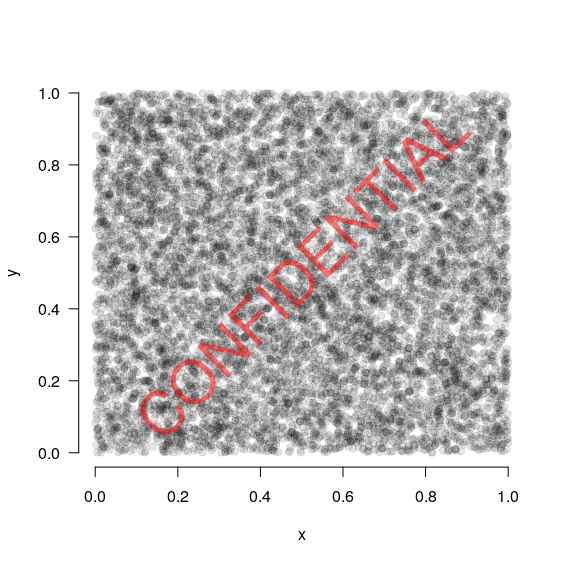
# Generate some example data
x <- runif(10000)
y <- runif(10000)
# Plot the points
plot(x, y, las=1, bty="n", pch=19, col=rgb(0,0,0, 0.1))
# Add a watermark
watermark("CONFIDENTIAL", col="red")
# Create an example dataset
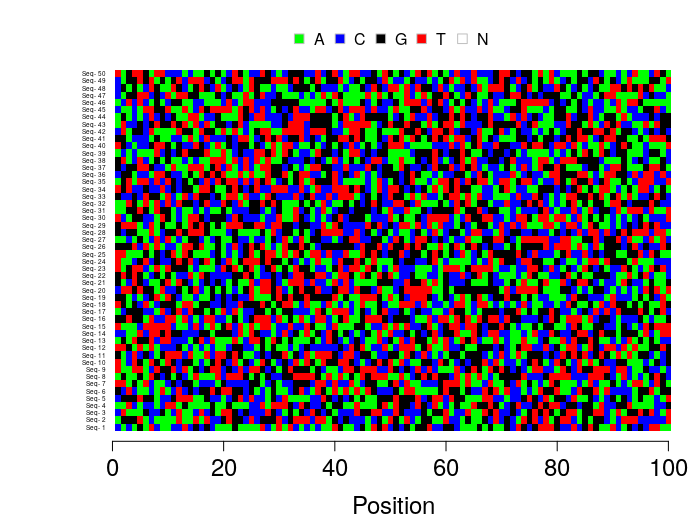
nSequences <- 50
nSites <- 100
alignment <- matrix(sample(c("A", "C", "G", "T"), nSequences*nSites, replace=TRUE),
nrow=nSequences, ncol=nSites)
rownames(alignment) <- paste("Seq-", 1:nSequences)
# Plot the FASTA
plotFASTA(alignment, xTicksCex = 1.5, xLabCex=1.5, sequenceLabelCex=0.4)
# Create some random points
n <- 50
coords <- data.frame(X=runif(n), Y=runif(n), Name="Test Label")
# Plot points and allow overlapping
plot(x=coords$X, y=coords$Y, bty="n", xaxt="n", yaxt="n", cex=3, xlab="X", ylab="Y")
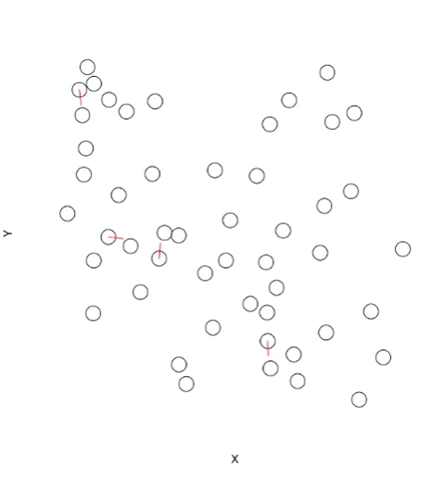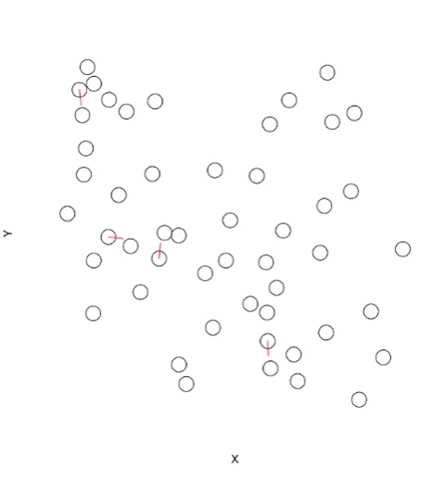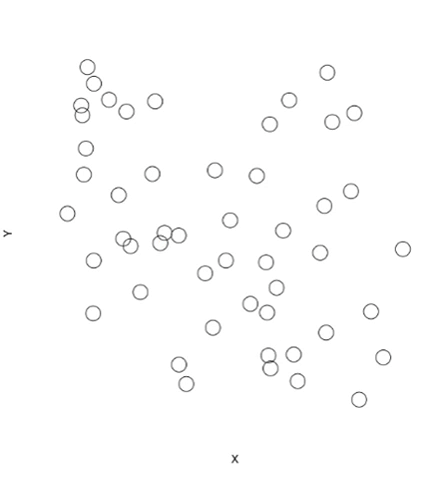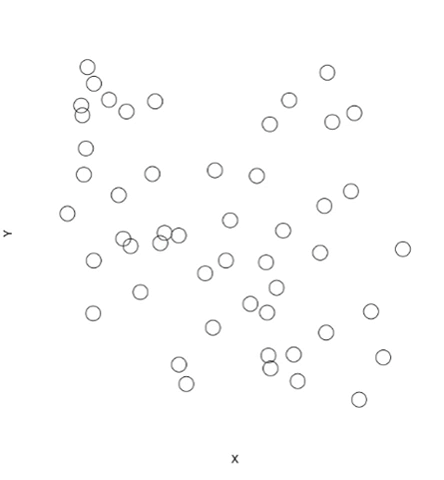
# Plot points and avoid overlapping
plot(x=NULL, y=NULL, xlim=range(coords$X), ylim=range(coords$Y), bty="n", xaxt="n", yaxt="n", xlab="X", ylab="Y")
addPoints(coords$X, coords$Y, cex=3, col.line="red")
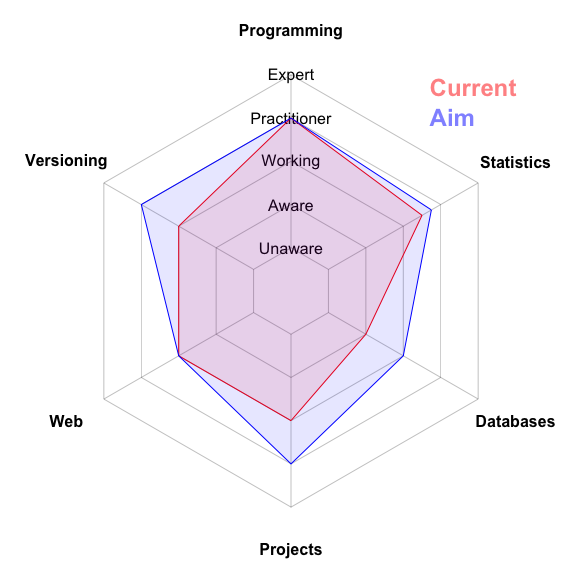
# Create a radar chart to map our current status for selected skills
radarChart(scores=c(4,3.5,2,3,3,3),
names=c("Programming", "Statistics", "Databases", "Projects",
"Web", "Versioning"),
levels=c("Unaware","Aware","Working","Practitioner", "Expert"))
# Add to the radar chart where we aim to get to
radarChart(scores=c(4,3.75,3,4,3,4),
names=c("Programming", "Statistics", "Databases", "Projects",
"Web", "Versioning"),
levels=c("Unaware","Aware","Working","Practitioner", "Expert"),
polygon.col="blue", add=TRUE)
# Add a legend
legend("topright",
legend=c("Current", "Aim"),
text.col=c(rgb(1,0,0, 0.5), rgb(0,0,1, 0.5)),
bty="n", text.font=2, cex=1.5, xpd=TRUE)