Sleep analysis with R.
SleepR latest version can be directly installed from Github using the devtools package.
devtools::install_github("boupetch/sleepr")
In SleepR, write_mdf() and read_mdf() functions are used to write and read records on disk. Files are converted from the European Data Format (EDF) to Morpheo Data Format1 (MDF). MDF is a simple, efficient and interoperable file format for biological timeseries. The format supports raw signal and metadata storage. MDF uses binary files for signals and JSON for metadata.
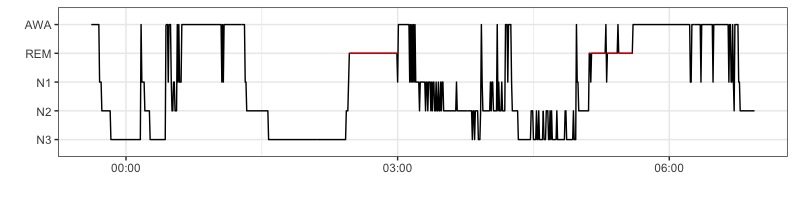
plot_hypnogram() function plots a hypnogram using ggplot2, higlighting REM sleep in red. The function takes as argument a dataframe containing labels, start and end times of events. AASM stages2 with labels AWA,REM,N1,N2,N3 by default. Labels can be redefined using the labels argument.
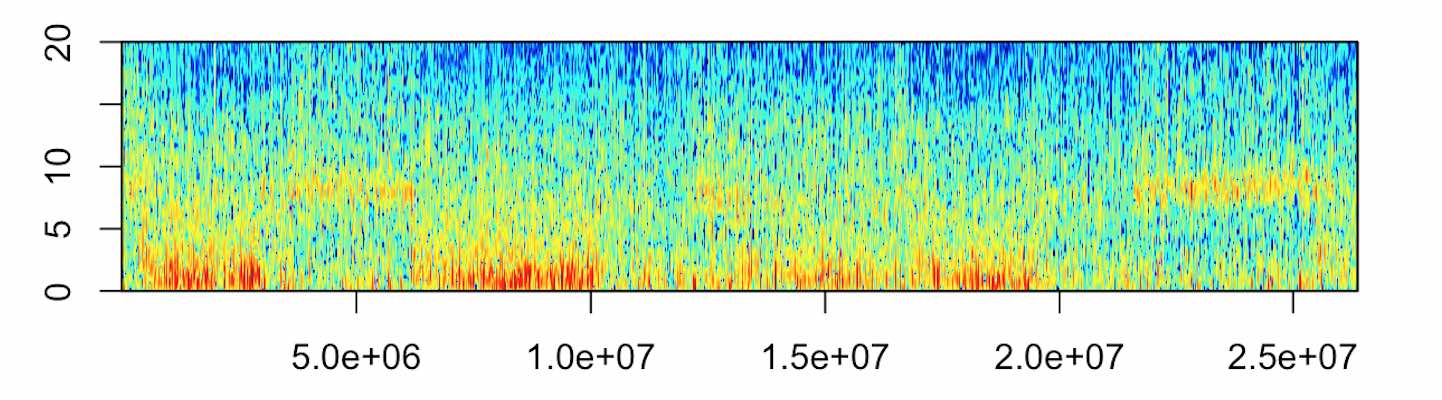
A spectrogram visually represents the frenquencies spectrum of a signal. Spectrogram are widely used to visualize and analyse EEG. plot_spectrogram() conveniently wrappers phonTools library spectrogram function to plot spectrograms.
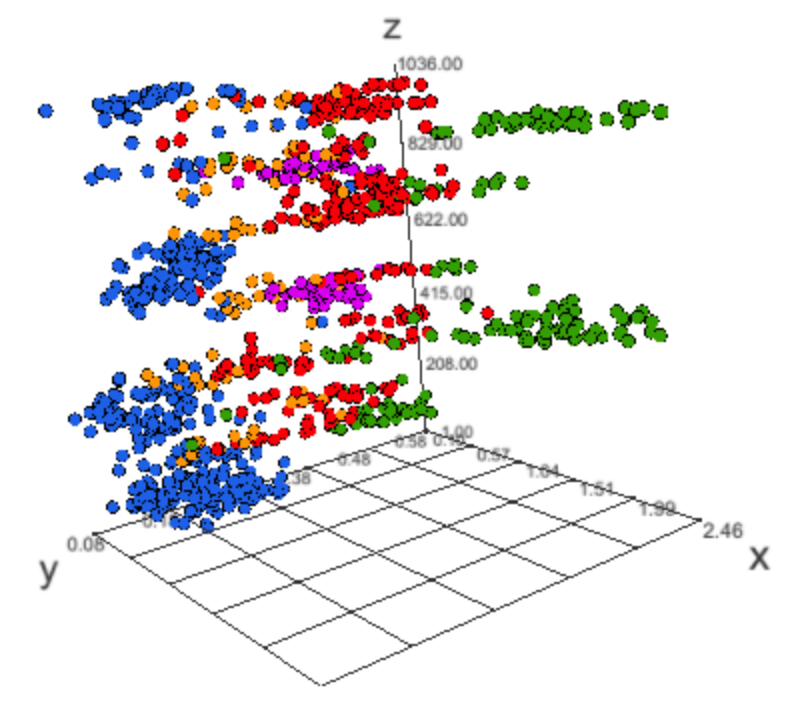
record <- read_mdf("mdf/isruc-1-89-1/")
powers <- sleepr::hypnogram_band_powers(record = record,
channel = "C3-M2")
powers$colors <- NA
powers$colors[powers$stage == "AWA"] <- "#1D60E7"
powers$colors[powers$stage == "REM"] <- "#D900F0"
powers$colors[powers$stage == "N1"] <- "#FD9509"
powers$colors[powers$stage == "N2"] <- "#F70007"
powers$colors[powers$stage == "N3"] <- "#339F03"
threejs::scatterplot3js(powers$delta, powers$theta,
as.integer(powers$epoch), size = 0.25,
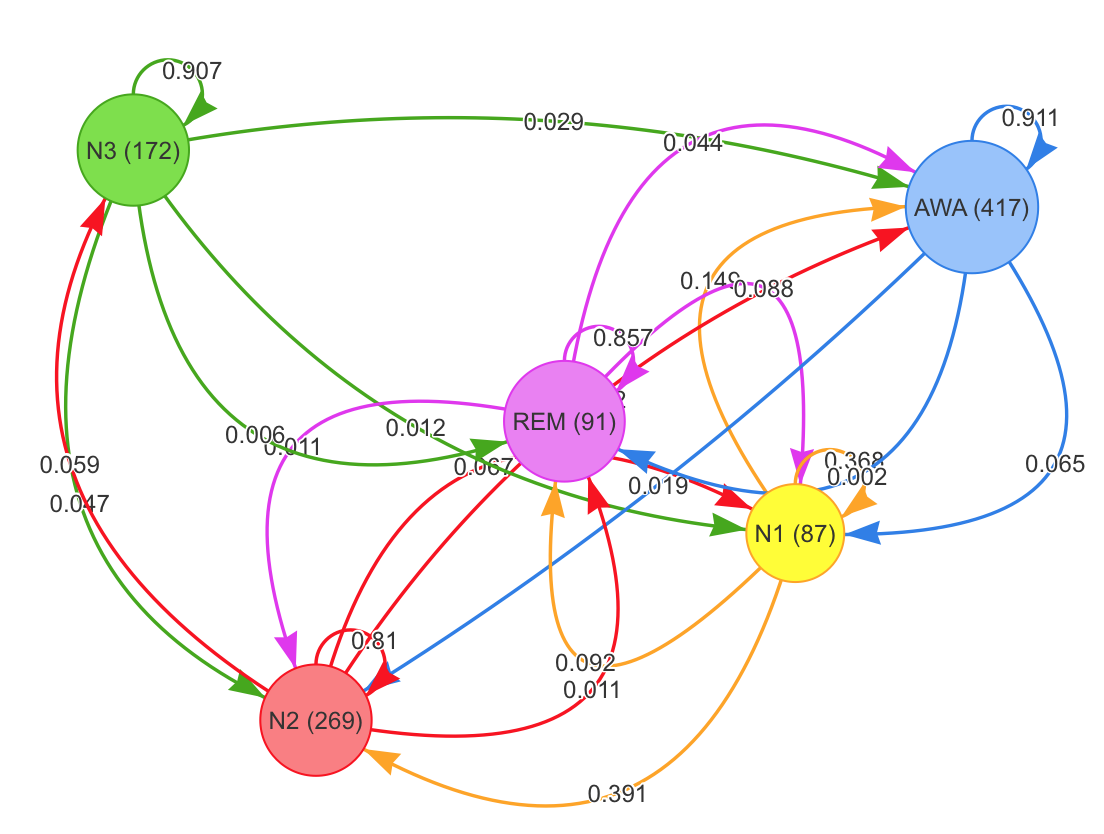
color= powers$colors)h <- hypnogram(read_mdf("mdf/1/89/",channels = NA)[["events"]])
tm <- transition_matrix(h)
knitr::kable(round(tm, digits=3))| AWA | N1 | N2 | N3 | REM | |
|---|---|---|---|---|---|
| AWA | 0.911 | 0.065 | 0.019 | 0.000 | 0.002 |
| N1 | 0.149 | 0.368 | 0.391 | 0.000 | 0.092 |
| N2 | 0.052 | 0.067 | 0.810 | 0.059 | 0.011 |
| N3 | 0.029 | 0.012 | 0.047 | 0.907 | 0.006 |
| REM | 0.044 | 0.088 | 0.011 | 0.000 | 0.857 |
h <- hypnogram(read_mdf("mdf/1/89/",channels = NA)[["events"]])
sleepr::plot_transitions(h)Various statistics can be computed from the polysomnographic data, signals and events. Durations are expressed in minutes, indexes in hours.
compute_all_stats() function call all the following functions over a vector of records paths. It returns a dataframe with a line for each record. EEG bands can be specified at this point to the bands argument.
# Write record to disk from EDF to MDF
write_mdf("path/to/edf", "path/to/mdf")
# Compute all stats from the record
compute_all_stats("path/to/mdf", bands = list(
delta = c(0.5,3.5), theta = c(3.5,8), alpha = c(8,12),
beta = c(12,30), gamma1 = c(30,40), gamma2 = c(40,60)))These functions compute statistics based on stage scoring.
rem_duration: Total duration of REM sleep in minutes.n1_duration: Total duration of N1 sleep in minutes.n2_duration: Total duration of N2 sleep in minutes.n3_duration: Total duration of N3 sleep in minutes.awa_duration: Total duration of wake in minutes.tts: Time To Sleep (N1+N2+N3+REM durations) in minutes.rem_tts: REM over TTS duration ratio.n3_tts: N3 over TTS duration ratio.n2_tts: N2 over TTS duration ratio.n1_tts: N1 over TTS duration ratio.tsp: Total Sleep Period.sleep_efficiency: Sleep Efficiency.sleep_latency: Sleep Latency.rem_latency: REM Sleep Latency.waso: Wake After Sleep Onset.
tts_pos_back: TTS duration in back position in minutes.tts_pos_back_pct: TTS duration in back position over TTS duration.tts_pos_left: TTS duration in left position in minutes.tts_pos_left_pct: TTS duration in left position over TTS duration.tts_pos_right: TTS duration in right position in minutes.tts_pos_right_pct: TTS duration in right position over TTS duration.tts_pos_stomach: TTS duration in left position in minutes.tts_pos_stomach_pct: TTS duration in stomach position over TTS duration.tts_pos_nonback: TTS duration in non-back position in minutes.tts_pos_nonback_pct: TTS duration in non-back position over TTS duration.
snoring_count: Snorings count.snoring_index: Snoring count per hour.snoring_duration: Total duration of snorings.snoring_duration_pct: Total duration of snorings over TTS duration.
ah_count: Apnea and hypopnea count.ah_hour: Apnea and hypopnea index by hour.ah_back: Apnea and hypopnea count in back position.ah_nonback: Apnea and hypopnea count in non-back position.ah_rem: Apnea and hypopnea count during REM stages.ah_nonrem: Apnea and hypopnea count during NREM stages.
ma_count: Micro-arousals count.ma_index: Micro-arousals index by hour.ma_duration: Total micro-arousals duration.ma_n1_duration: Total micro-arousals duration during N1.ma_n2_duration: Total micro-arousals duration during N2.ma_n3_duration: Total micro-arousals duration during N3.ma_rem_duration: Total micro-arousals duration during REM.ma_n1_count: Micro-arousals count during N1.ma_n2_count: Micro-arousals count during N2.ma_n3_count: Micro-arousals count during N3.ma_rem_count: Micro-arousals count during REM.ma_n1_index: Micro-arousals index during N1.ma_n2_index: Micro-arousals index during N2.ma_n3_index: Micro-arousals index during N3.ma_rem_index: Micro-arousals index during REM.
rem_count: Count of Rapid-Eye-Movements.rem_index: Rapid-Eye-Movements index by hour.rem_avg_duration: Rapid-Eye-Movements average duration.
cycles_classic_count: Regular cycles count.cycles_begin_count: Starting cycles count.cycles_end_count: Ending cycles count.cycles_rem_count: REM cycles count.cycles_classic_duration: Regular cycles total duration.cycles_begin_duration: Starting cycles total duration.cycles_rem_duration: REM cycles total duration.cycles_end_duration: Ending cycles total duration.cycles_classic_avg_duration: Regular cycles average duration.cycles_begin_avg_duration: Starting cycles average duration.cycles_rem_avg_duration: REM cycles average duration.cycles_end_avg_duration: Ending cycles average duration.
SleepR uses the following nomenclature for annotated events labels:
AWA: Awake stage.REM: Rapid Eye Movement (REM) stage.N1: Non-REM (NREM) stage 1.N2: Non-REM (NREM) stage 2.N3: Non-REM (NREM) stage 2.
cycle-CLASSICA cycle which roughly follows the pattern: N1-N2-N3-R.cycle-BNThe beginning of the night; a cycle which roughly follows the pattern: N1-N2-N3-W/1 (without REM).cycle-ENThe end of the night; a cycle which roughly follows the pattern: N1-N2-R.cycle-REMA REM cycle.
rightleftbackstomach
Rapidemicro-arousal
SleepR can download open databases with simple function calls.
A collection of sleep records from the [Institute for Systems and Robotics (ISR)](Institute for Systems and Robotics (ISR)) of the University of Coimbra, Portugal.
download_isruc("./")61 polysomnograms hosted on Physionet5.
download_sleepedfx("./")
20 whole-night PSG recordings coming from healthy subjects.
download_dreams_subjects("./")
A collection of 108 polysomnographic recordings registered at the Sleep Disorders Center of the Ospedale Maggiore of Parma, Italy and hosted on Physionet5.
download_capslpdb("./")
Sleepviewer.com is an online application to visualize sleep records from various open data sources. It uses SleepR to manage and process records.
Testing use testthat.
R CMD Rd2pdf . && mv ..pdf sleepr.pdf && rm -r .Rd2pdf*
-
P. Bouchequet, D. Jin, G. Solelhac, M. Chennaoui, D. Leger, «Morpheo Data Format (MDF), un nouveau format de données simple, robuste et performant pour stocker et analyser les enregistrements de sommeil», Médecine du Sommeil, vol. 15, n 1, p. 48‑49, march 2018.
-
R.B. Berry, R. Brooks, C.E. Gamaldo, S.M. Harding, C.L. Marcus, B.V. Vaughn, The AASM Manual for the Scoring of Sleep and Associated Events, 2013.
-
Khalighi Sirvan, Teresa Sousa, José Moutinho Santos, and Urbano Nunes. ISRUC-Sleep: A comprehensive public dataset for sleep researchers. Computer methods and programs in biomedicine 124 (2016): 180-192.
-
B Kemp, AH Zwinderman, B Tuk, HAC Kamphuisen, JJL Oberyé. Analysis of a sleep-dependent neuronal feedback loop: the slow-wave microcontinuity of the EEG. IEEE-BME 47(9):1185-1194 (2000).
-
Goldberger AL, Amaral LAN, Glass L, Hausdorff JM, Ivanov PCh, Mark RG, Mietus JE, Moody GB, Peng C-K, Stanley HE. PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation 101(23):e215-e220 Circulation Electronic Pages; 2000 (June 13).
-
S. Devuyst, T. Dutoit, P. Stenuit, M. Kerkhofs, E. Stanus, "Canceling ECG Artifacts in EEG using a Modified Independent Component Analysis Approach", EURASIP Journal on Advances in Signal Processing, Volume 2008, Article ID 747325, Accepted 31 July 2008.
-
MG Terzano, L Parrino, A Sherieri, R Chervin, S Chokroverty, C Guilleminault, M Hirshkowitz, M Mahowald, H Moldofsky, A Rosa, R Thomas, A Walters. Atlas, rules, and recording techniques for the scoring of cyclic alternating pattern (CAP) in human sleep. Sleep Med 2001 Nov; 2(6):537-553.