tshidoshi
nevhungwili - very cool name!!
both a local edit and online edit
https://docs.google.com/document/d/18mzlAB3q7SBzJoVpUcthZDuCM0NsvL9fw_FUs-4fud4/edit?usp=sharing
Testing structure for tracking/organizing scripts that create plots/tables for publications:
-
All data in a single 'data' folder (or loaded directly by scripts in this repo from another online repository)
-
One folder for each small script that produces a plot and/or table (keeping separate analyses separate as much as possible), with outputs from each script kept in the same folder.
-
This README file to show the figures (and links to tables) intended to go in a paper, ordered by paper section, and including the intended Title and Caption (click on the dropdown at the topleft to get a quick overview via the TOC).
Editing the publication draft 'body' text happens in an appropriate platform (e.g. using track changes in Word), but only links to figures/tables in GitHub1 are included in the draft while analysis/formatting is ongoing (i.e. avoid pasting a figure/table into the text if it might need to be changed later).
When analysis is complete, and plots/tables are in their final form, they can then be incorporated with main text2.
Key thing: every plot/table is kept with the script that made it, so it can be inspected/corrected easily if need be (click on the link/figure, and then open the containing folder to see - and comment on - the script that generated it); no time wasted trying to paste every new version of a plot into the Word doc (and probably making mistakes with versions).
Should include details about the paper here - e.g. the running title, where to find the draft, a basic summary, etc. .. when the paper is published, this can be updated, and the repo can be made public (if desired) to share the code used in the paper.
Some basic information about the data used should go here.
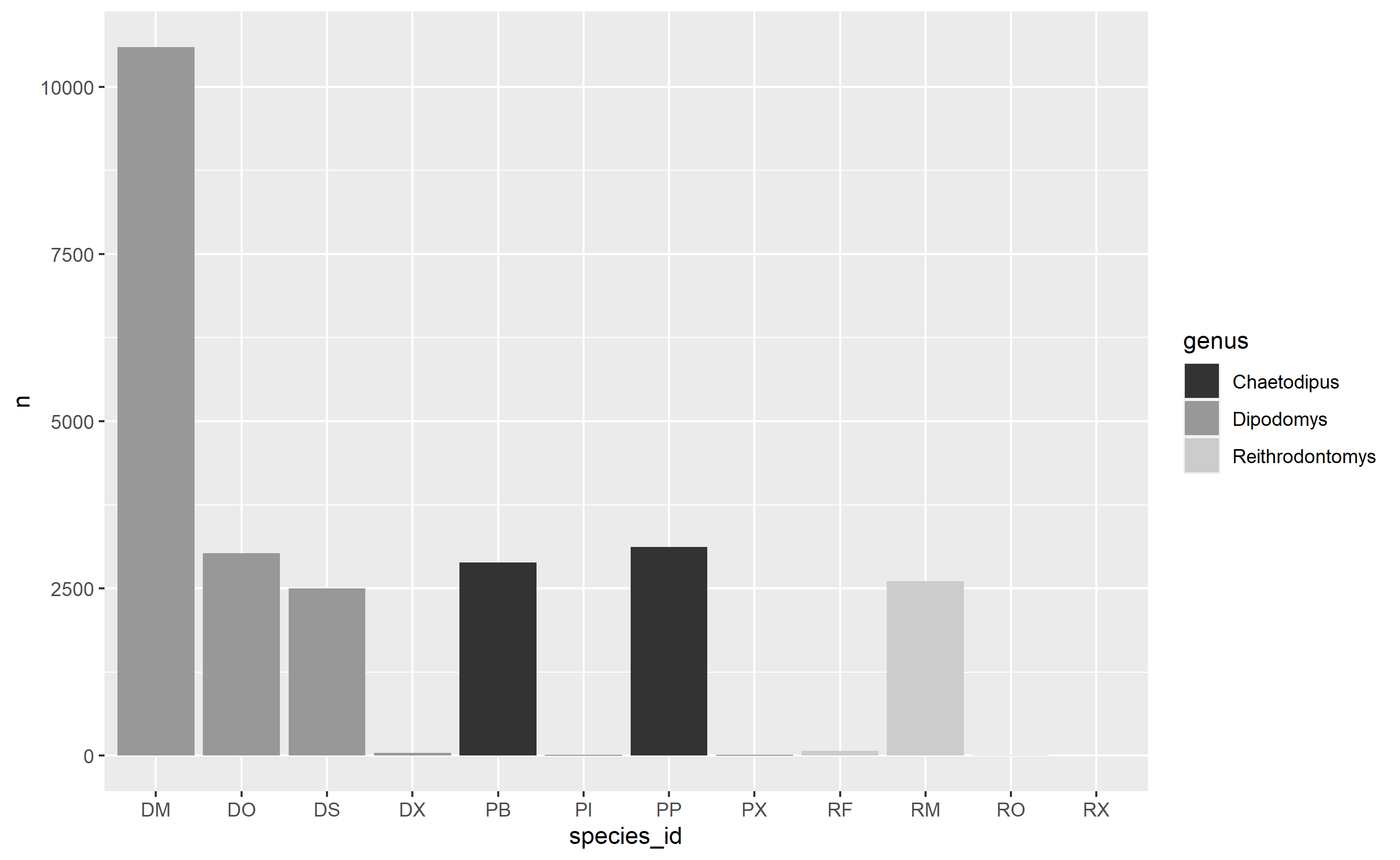
Title: Counts of species observed in the three most diverse genera
Title: Relative counts of species observed in three most diverse genera
Caption: Species counts were conducted as per the methodology described in the Methods section. Genera with more than 3 observed species were considered 'diverse', with relative counts shown here.
Footnotes
-
To get the link to a subheading in this README, move the mouse to the left of the subheading and right-click on the chain-link icon that appears, and then select 'Copy link address'. To link directly to the figure/table, click on the appropriate figure/table link in this README (or navigate to its position in the repo), and then copy the URL from the navigation bar. ↩
-
To transfer tables into Word in preparation for publication, open the csv file in Notepad (if on your computer, else look for the 'Raw' button in GitHub), copy and paste the contents into Word, and do the following: select the text that you want to convert, and then click Insert > Table > Convert Text to Table; in the Convert Text to Table box, choose the options you want. Under Table size, make sure the numbers match the numbers of columns and rows you want. ↩