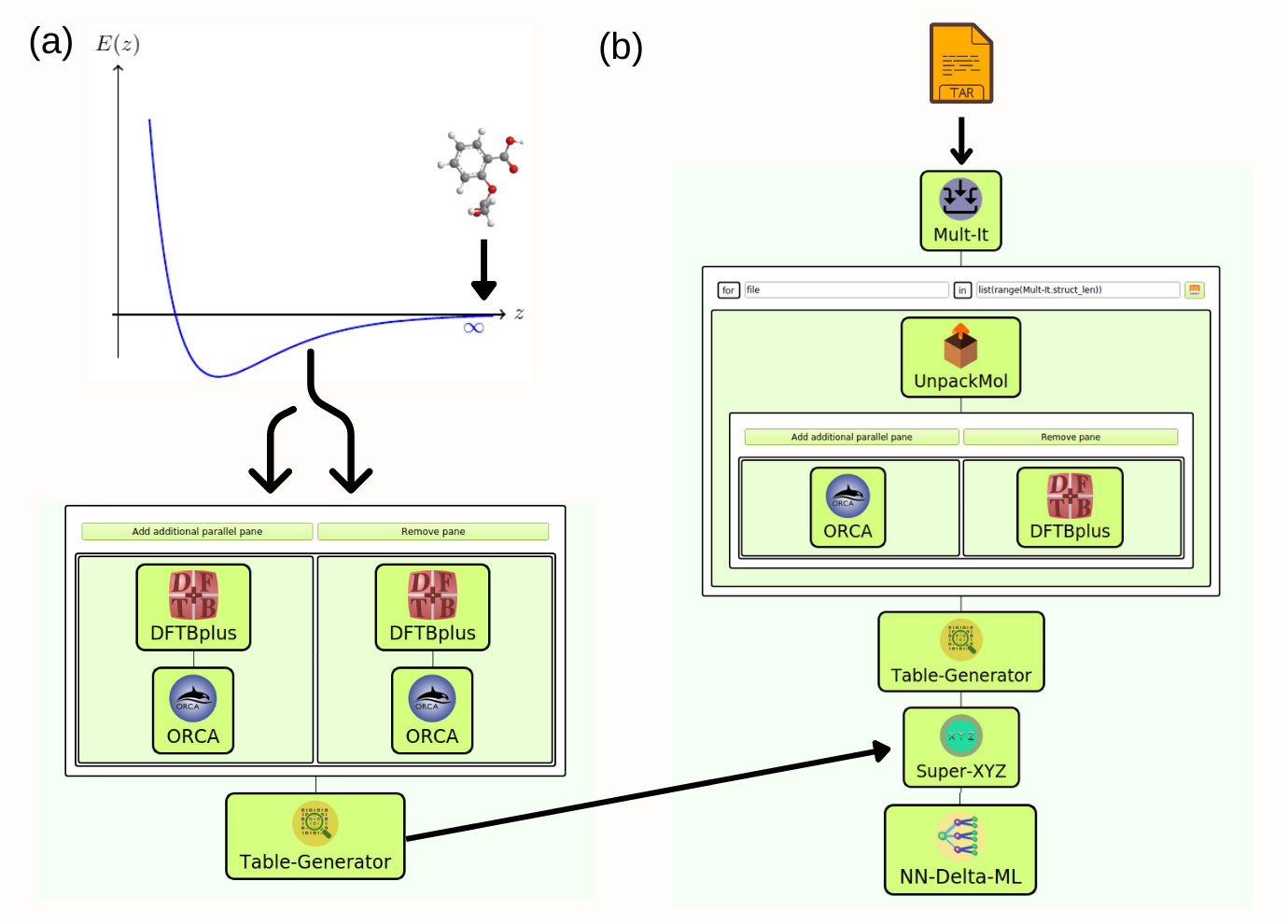
This workflow developed in the SimStack framework enables the correction of DFTB potential energy surfaces into ab-initio methods using the
In the folder WaNos there are several different WaNos: DFT-Turbomole, DFTBplus, Mult-It, NN-Delta-ML, ORCA, Super-XYZ, Table-Generator and UnpackMol, used to build the workflow. Below we describe each one and the main parameter exposed.
- Load a set of molecular trial structures in a
.tarfile. - Unpack all molecular structures inside the AdvancedForEach loop control in the Simstack framework.
- Compute the reference energy of the system.
- Run the DFT calculations using ORCA or Turbomole codes, ORCA or DFT-Turbomole WaNos.
- Run the DFTB calculations using BFTB+ code using DFTBplus WaNo.
- Arrange all the total energy values of the system in a table format (Table-Generator).
- Append all files from the
.tarinput file in a specific order and shift the total DFT and DFTB energies from the previously computed reference energies. - Compute the
$\Delta$ energy to generate the machine learning (ML) model and the learning report. - Apply the ML model to predict the
$\Delta E$ for a similar system when stimulated via the DFTB method.
Fig 1 This workflow aims to create an ML model to correct DFTB method accuracy concerning the DFT level. It is composed of DFT-Turbomole, DFTBplus, Mult-It, NN-Delta-ML, ORCA, Super-XYZ, Table-Generator and UnpackMol WaNos connected by the AdvancedFor loop control. (a) When the system is far from the equilibrium region, we compute the reference energy for DFT and DFTB levels. (b) In this step, the set of molecular structures in a .tar file is loaded, and a high throughput calculation (single shot) is performed for DFT and DFTB theory levels. The workflow automatically creates a machine learning report at the end of ML model generation.
Here you will find the steps to install DFTB+ code and python dependencies necessary to run the workflow. Of course, we assume that you already installed the DFT code. You can choose any option available in the list of quantum chemistry codes. In this case, we use ORCA or Turbomole.
Install conda environment, the following packages would be needed:
conda create --name environment_name python=3.6 --file environment.yml
Install via pip ordered-enum
pip3 install ordered_enum
⚠️ If you are installing DFTB+ on a different machine: Be very careful and make the necessary adjustments.
- git clone -b machine-learning https://github.com/tomaskubar/dftbplus.git
- cd dftbplus
- module load gnu8/8.3.0
- module load openblas/0.3.7
- module load cmake
- mkdir _build
- FC=gfortran CC=gcc cmake -DCMAKE_INSTALL_PREFIX=$HOME/opt/dftb+ -B _build .
- cmake --build _build -- -j
- cmake --install _build
+ Check if the `dftb+` executable exist in the dftbplus/_build/prog/dftb+/ folder. If so, then everything is okay. + be cautious with conda env., it must have libgfortran5 if the installation wants to be done inside the conda environment- Molecular geometry far from the equilibrium region (as shown in Fig 1 (a)).
- Set of randmized structures around the Potential energy surface (
.tarfile as shown in Fig 1 (b)).
- Machine learning report
- Pickle files of the machine learning model.
In the video
- J. Behler and M. Parrinello, Physical Review Letters 98, 146401 (2007).
- J. Zhu, V. Q. Vuong, B. G. Sumpter, and S. Irle, MRS Communications 9, 867 (2019).
- R. Ramakrishnan, P. O. Dral, M. Rupp, and O. A. von Lilienfeld, Journal of Chemical Theory and Computation 11, 2087 (2015).
- C. R. Rêgo, J. Schaarschmidt, T. Schlöder, M. Penaloza-Amion, S. Bag, T. Neumann, T. Strunk, and W. Wenzel, Frontiers in Materials , 283.
- C. L. Gómez-Flores, D. Maag, M. Kansari, V.Q. Vuong, S. Irle, F. Gräter, T. Kubař, and M. Elstner, Journal of Chemical Theory and Computation 18, 1213 (2022).