You can find our paper on arXiv 📄.
Authors: Patrick Knab, Katharina Prasse, Sascha Marton, Christian Bartelt, and Margret Keuper
This repository is organized into two main components:
- Concept Extraction
- GCBM Training and Visualization
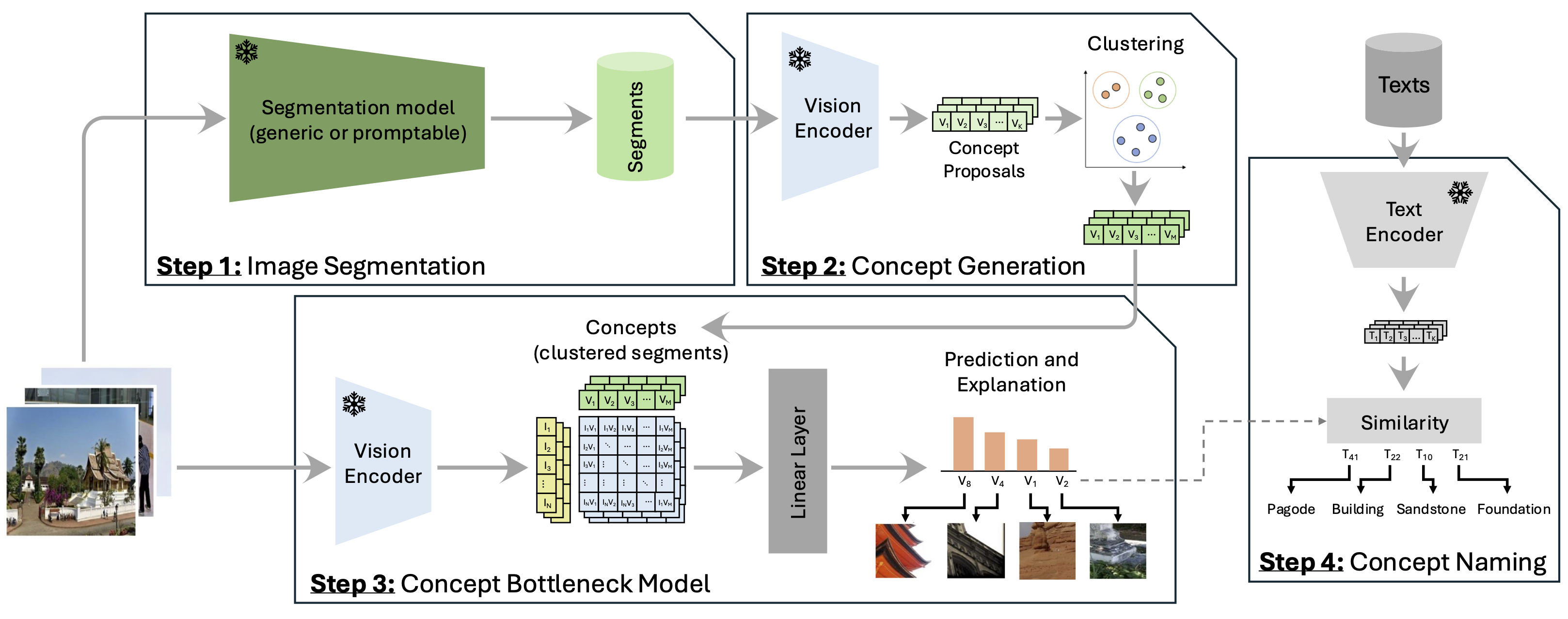
Choose segmentation models based on your computational budget and the latest advancements. GDINO is the fastest model in our evaluation. We provide .yml files for all segmentation methods used in this work.
Build SAM2, GroundingDINO, and Semantic SAM from source, as suggested in their respective GitHub repositories. Place the repositories in the utilities folder and adjust the paths as necessary.
Execute the segmentation method by running the respective script in the Segments folder:
python scripts/create_segments_SAM2.py --dataset cifar100 --device cuda:0Embed the segments into the selected embedding space by running the corresponding script in the Segments folder. We primarily report on CLIP models [RN-50, ViT-B16, ViT-L14] and save embeddings to reuse in GCBM training for efficiency.
python scripts/emb_segments_CLIP.py --dataset cifar100 --device cuda:0 --emb CLIP-RN50 --seg_method SAM2For datasets without a provided validation set, create one by splitting the training set. Following common practices, we evaluate on the ImageNet-Val split, so we need a new set for hyperparameter validation.
Split the training set into training and validation sets (90:10). Ensure the test set is named test, as some datasets have train and val sets.
Adjust the dataset path in the script to your existing training set and the new validation set.
python data/Datasets/scripts/create_val.pyEmbed images to reuse in GCBM training for time and resource efficiency.
python data/Datasets/scripts/embed_CLIP.pyWith all necessary files correctly stored in the data directory, initiate GCBM training:
python gcbm_training/gcbm_testing.pyNecessary Files in data/:
classes/
Contains class information for the dataset (required for new datasets).Concepts/
Directory to store previously extracted concepts.Datasets/
Contains the training, testing, and validation datasets.Embeddings/- Subset of embeddings (if not segmenting all training images).
- Embedded files for test, validation, and training sets.
Segments/- Contains segmented images.
- Includes embeddings in the
Seg_embs/subdirectory.
We also provide multiple Jupyter notebooks in gcbm_training/ to facilitate training and hyperparameter tuning.
To apply the code to a new dataset, modify the relevant sections in utils/gcbm.py as indicated by the TODO comments.
Explore interactive explanations with GCBM across different datasets using the Jupyter notebooks in interpretation/.
Install the required dependencies using the provided requirements.txt file:
pip install -r requirements.txtThis environment supports the creation of CLIP embeddings. We have also created specialized environments for generating DINOv2 embeddings and using various segmentation models.
The repository is structured as follows:
.
├── concept_extraction/
├── gcbm_training/
├── data/
│ ├── classes/
│ ├── Concepts/
│ ├── Datasets/
│ ├── Embeddings/
│ ├── Segments/
├── experiments/
├── utils/
├── interpretation/
├── requirements.txt
└── README.mdconcept_extraction/: Scripts and modules for extracting concepts.gcbm_training/: Code for training the GCBM model.data/: Directories for classes, concepts, datasets, embeddings, and segments.experiments/: Code for experiments detailed in the main paper and supplementary material.utils/: Helper scripts for running experiments.interpretation/: Tools for visualizing GCBM explanations.
Important: Some folders in the data/ directory may be empty. Due to size limitations, we couldn't upload the complete dataset and corresponding concepts. Therefore, these folders lack content.
To run the scripts correctly, please add the required datasets before execution.
For more details, please refer to the sections and scripts within the repository or consult the paper.