A quantitative framework for evaluating data structure preservation by dimensionality reduction techniques
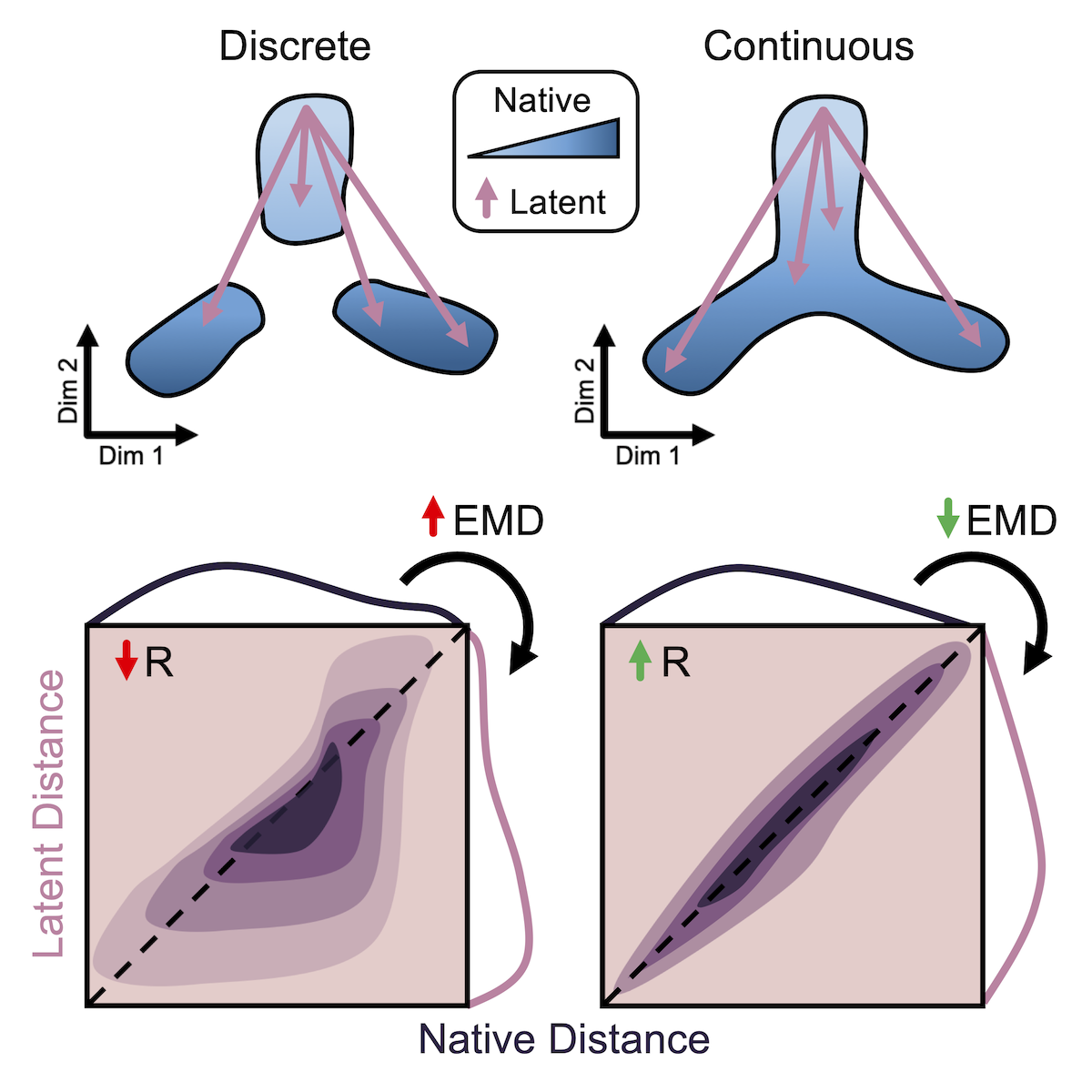
High-dimensional data is integral to modern systems biology. Computational methods for dimensionality reduction are being rapidly developed for applications to single-cell and multi-modal technologies. In order to understand what these nonlinear transformations do to the underlying biological patterns in our data, we developed a framework of quantitative metrics for global and local distance preservation. See our Cell Reports publication for details and analysis of existing dimensionality reduction methods.
Contain utility functions for manipulating datasets and comparing feature-reduced latent spaces.
Documentation available at Ken-Lau-Lab.github.io/DR-structure-preservation/.
Consult distance_preservation_tutorial.ipynb for info on how to perform global and local structure preservation analyses on low-dimensional embeddings of your own datasets.
Install using pip:
pip install -r requirements.txtIn order to use the "FIt-SNE" implementation of t-SNE, you'll need to download FFTW and compile the code from the FIt-SNE repo.
Clone the scvis and ZIFA packages and install with python setup.py install from their home directories.