drep 
Graphical and Numerical Representation of DNA Sequences
R library for characterizing biological sequences graphically and numerically. If you would like to get the gist of implemented approach please read the following article
Bo Liao, Kequan Ding, A 3D graphical representation of DNA sequences and its application. Volume 358, Issue 1, 2006, Pages 56-64.
Install
In order to install and load the package run the following code in the R console
library(devtools)
install_github('officialprofile/DynamicRepresentation')
library(drep)How to use it
In most cases a single line of code can yield satisfactory results. For example
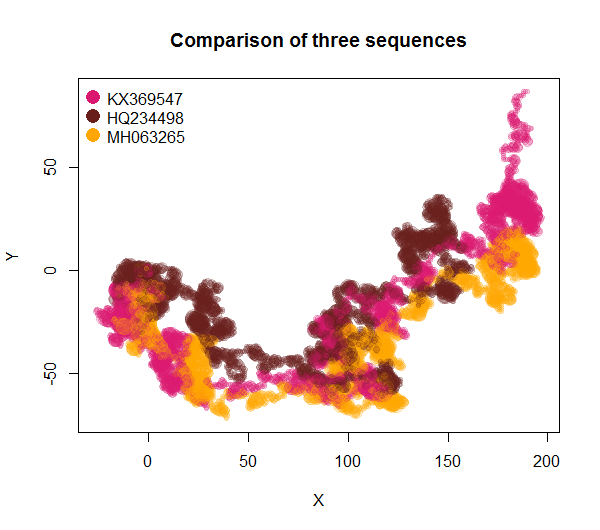
plot2DGraph(c('KX369547', 'HQ234498', 'MH063265'), genbank = TRUE, main = 'Comparison of three sequences')returns a ready-to-use graph of the ZIKV genomes
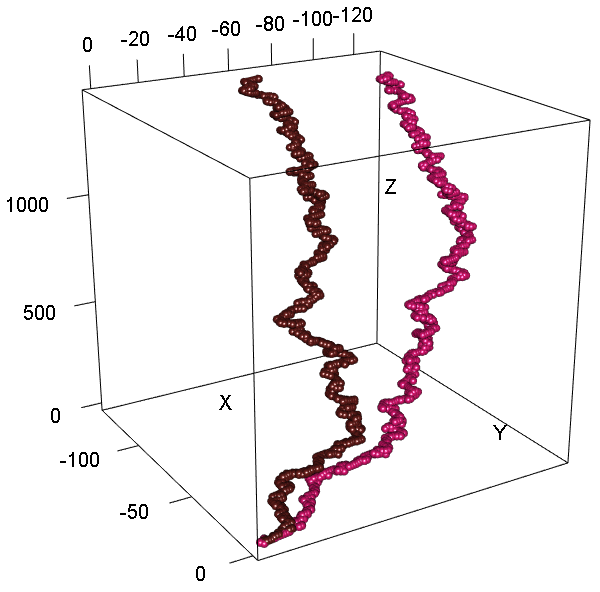
For three-dimensional representation one can use plot3DGraph. The function is based on rgl library.
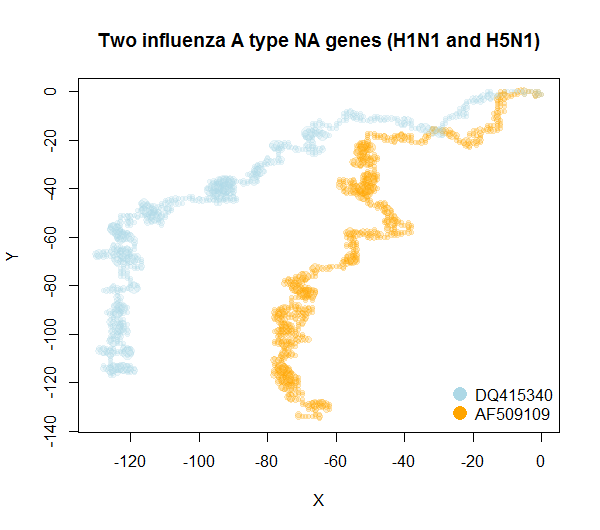
plot3DGraph(c('DQ415340','AF509109'), genbank = TRUE, radius = 10)plot2DGraph(c('DQ415340','AF509109'), genbank = TRUE,
legend.pos = 'bottomright',
colorset = c('lightblue', 'orange'),
main = 'Two influenza A type NA genes (H1N1 and H5N1)')Notice that plotting such graphs can be achieved solely by putting the GenBank accession numbers. plot2DGraph as well as plot3DGraph are intended to be quite versatile though.
In similar fashion one can obtain numerical characteristics by employing dRep function, e.g.
dRep(c('KX369547', 'HQ234498'), genbank = TRUE)returns the following dataframe
| len | mi_x | mi_y | sqrt | ... | |
|---|---|---|---|---|---|
| KX369547 | 10769 | 84.660 | -16.061 | 86.170 | ... |
| HQ234498 | 10269 | 75.171 | -17.691 | 77.224 | ... |
Naturally, instead of using data from GenBank, one can apply implemented method to one's own sequence or vector of sequences, e.g.
seq <- 'ACCCTCGCGCCGCGATTCTACGGACCCTGAAAATG'
dRep(seq)