Kvik
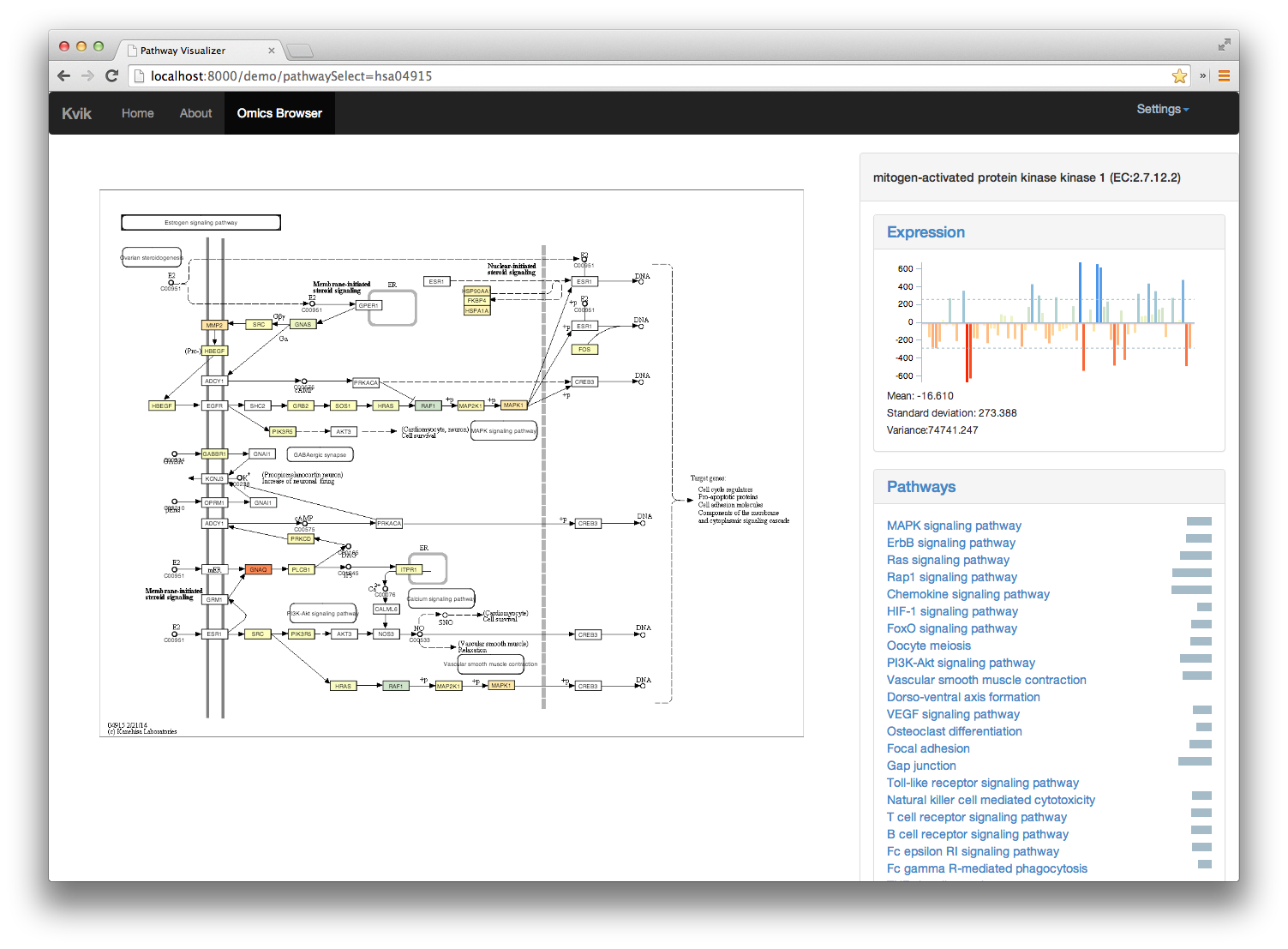
An interactive system for exploring the dynamics of carcinogenesis through integrated studies of biological pathways and genomic data. It provides researchers with a lightweight web application for navigating through biological pathways from the KEGG database integrated with genomic data from the NOWAC postgenome biobank.
About
My master's thesis project at the BDPS group at UiT. You can find the thesis here and presentation of the thesis here.
Source Code
The source code is found in the src directory. It consists of four individual components all written in Go. The Data Engine expects data from the NOWAC biobank and will not run without it. Note that both the Frontend and the Data Engine depends on gorest changeset 3ba1ba16240d and other libraries, but hopefully these are downloaded when you run go install.
Contact
Feel free to contact me at bjorn.fjukstad@uit.no!