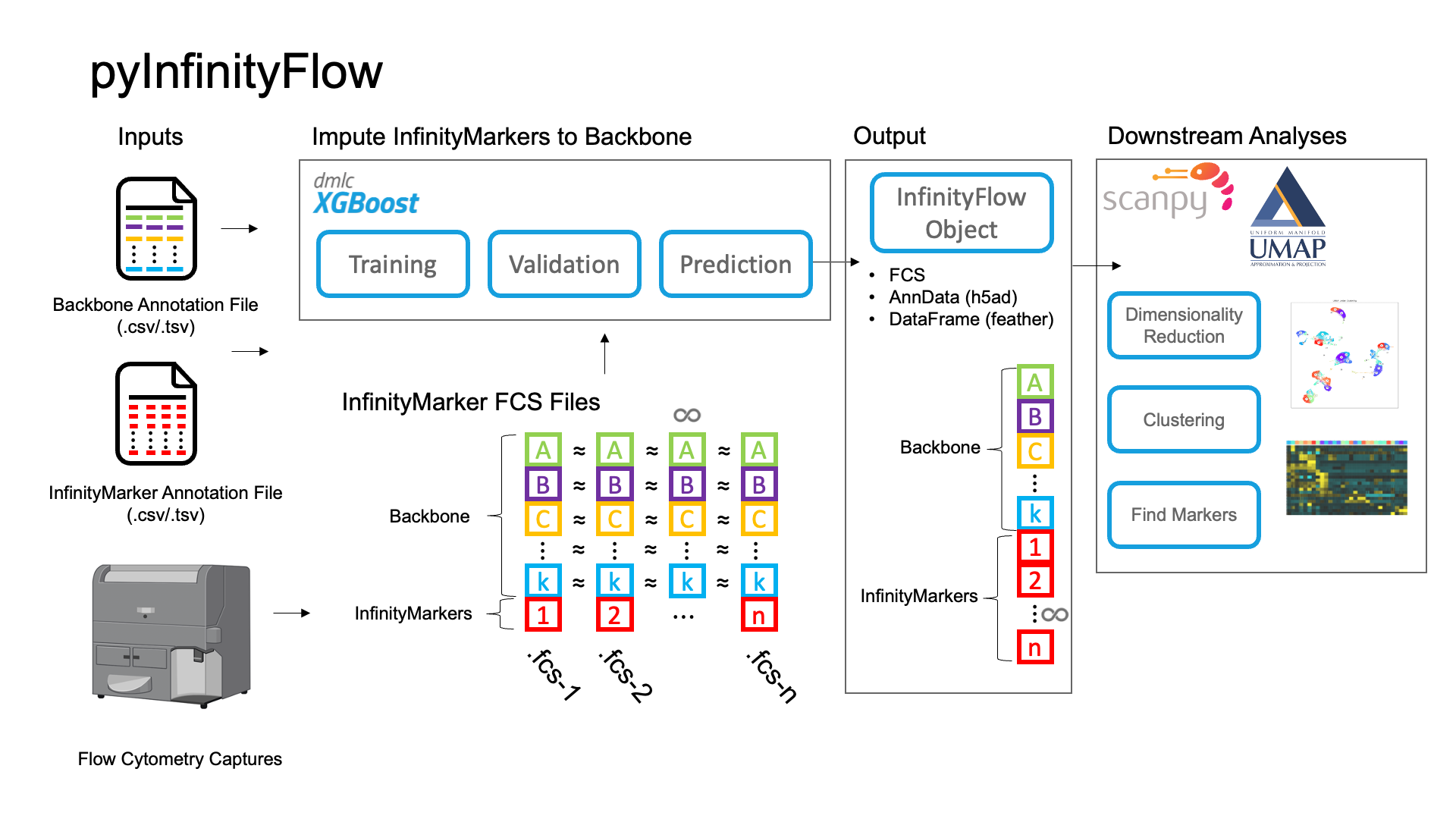
pyInfinityFlow is a Python package that enables imputation of hundreds of features from Flow Cytometry using XGBoost regression1. It is an adaptation of the original implementation in R2 with the goal of optimizing the workflow for large datasets by increasing the speed and memory efficiency of the analysis pipeline.
The package includes tools to read and write FCS files, following the FCS3.1 file standard, into AnnData objects, allowing for easy downstream analysis of single-cell data with Scanpy3 and UMAP4.
Read more about the pyInfinityFlow package on its Read the Docs page!
It is recommended to set up a virtual environment to install the package.
Creating a new conda environment and installing pyInfinityFlow:
conda create -n pyInfinityFlow python=3.8
conda activate pyInfinityFlow
pip install pyInfinityFlowThen pyInfinityFlow will be installed in a conda environment named 'pyInfinityFlow'.
To run the pyInfinityFlow pipeline, we can use this command:
pyInfinityFlow --data_dir /home/kyle/Documents/GitHub/pyInfinityFlow/example_data/mouse_lung_dataset_subset/ \
--out_dir /media/kyle_ssd1/example_outputs/ \
--backbone_annotation /home/kyle/Documents/GitHub/pyInfinityFlow/example_data/mouse_lung_dataset_subset_backbone_anno.csv \
--infinity_marker_annotation /home/kyle/Documents/GitHub/pyInfinityFlow/example_data/mouse_lung_dataset_subset_infinity_marker_anno.csv