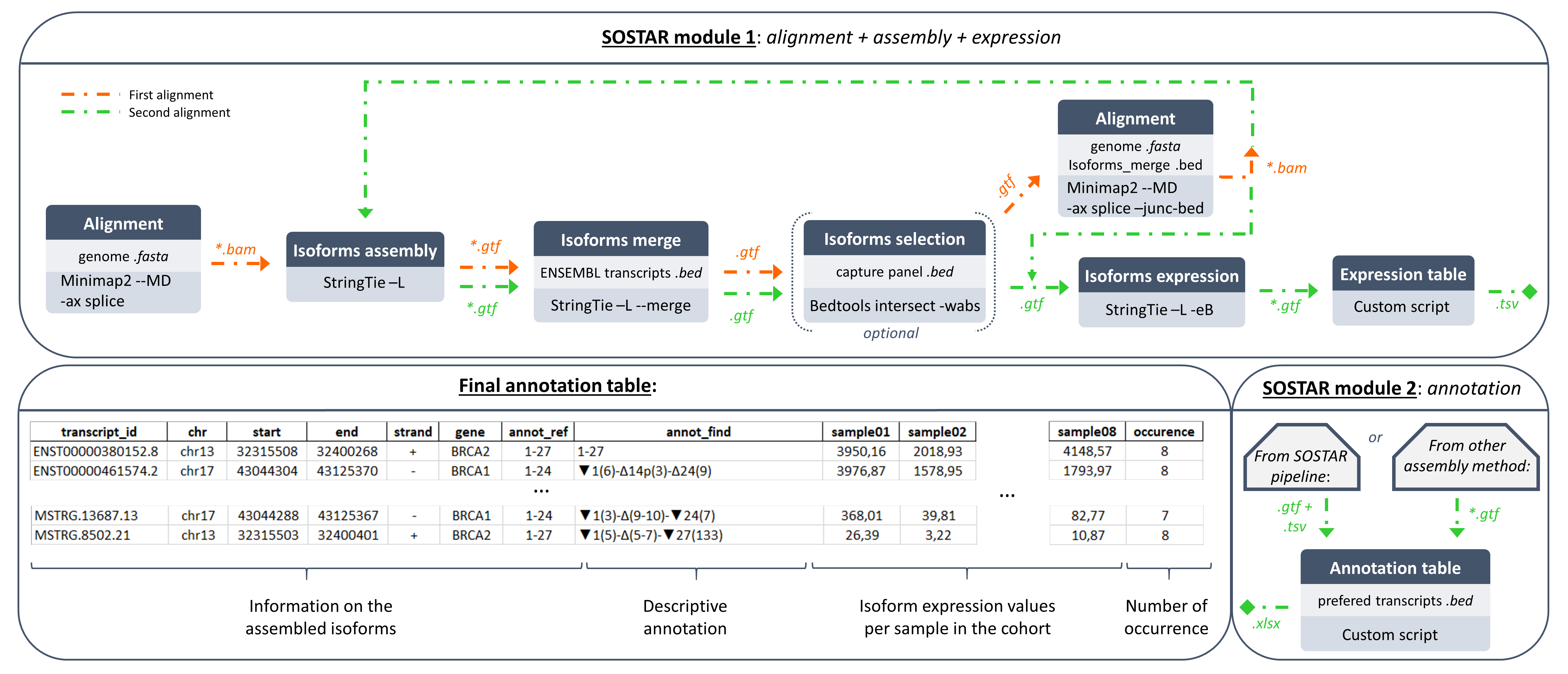
SOSTAR is a versatile pipeline to assemble and describe isoforms from long read sequencing using a new annotation tool. The pipeline is divided into two modules that can be run separately.
- First module: The first module performs alignment (in two rounds) using minimap2, assembled isoforms and computed isoform expression using StringTie.
- Second module: The second module provides a descriptive and comprehensive annotation of each assembled isoform. Isoforms are described relative to reference transcripts (provided by user) by an annotation including only alternative splicing events.
This pipeline is a Snakemake workflow.
Dependencies:
- Snakemake : 7.32.4 or greater.
- Singularity : 3.5.3 or greater.
The workflow automatically uses a docker image which contains the other tools required.
Download the Snakefile and rules of SOSTAR pipeline:
git clone https://github.com/LBGC-CFB/SOSTAR.git
cd ./SOSTAR
To configure this workflow, modify the
./config/config.yamlfile according to your needs.
-
genome: Fasta file of reference genome. Example in./tests/hg19_chr17.fa. -
ensembl_annot: GTF annotation file of known reference transcrits described in databases (Ensembl, Refseq, ...). Example in./tests/gencode.v19.annotation_chr17.gtf. -
transcripts_list: Txt file of the reference transcripts used for the annotation step. Name must match the "transcript_id" attribute from theensembl_annotfile. Example in./tests/reference_transcripts_chr17.txt. -
indir: Path of the directory containing the.fastq.gzinput files. -
outdir: Path of the outdirectory. -
samples: names of the differents samples. -
threads: number of threads to use. -
bedtools: option to indicate whether or not bedtools will be used to filter out transcripts from the genes specified in the reference transcript file. "True" or "False".
Given that the workflow has been properly configured, it can be executed as follows:
cd ./workflow
snakemake --use-singularity --cores
SOSTAR/outdir
├── alignment
│ └── aligned
│ ├── {sample}.aligned.bam
| ├── {sample}.aligned.bam.bai
│ └── ...
│ └── realigned
│ ├── {sample}.realigned.bam
| ├── {sample}.realigned.bam.bai
│ └── ...
├── assembly
│ └── aligned
│ ├── {sample}.assembly.aligned.gtf
│ └── ...
│ └── realigned
│ ├── {sample}.assembly.realigned.gtf
│ └── ...
│ ├── transcripts.merged.aligned.filter.bed
│ ├── transcripts.merged.aligned.filter.gtf
│ └── transcripts.merged.realigned.filter.gtf
├── expression
│ ├── {sample}.expression.gtf
│ └── ...
├── ref_transcripts_annotation.bed
├── ref_transcripts_annotation.gtf
└── SOSTAR_annotation_table_results.xlsx
alignmentfolder: contains subfolders of all aligned and sorted {sample} <.bam> with their corresponding index <.bai> either on the first (aligned subfolder) or the second (realigned subfolder) round of alignment.assemblyfolder: contains subfolders of all assembled <.gtf> {sample} with StringTie either on the first (aligned subfolder) or the second (realigned subfolder) round of alignment plus the global <.gtf> and it corresponding <.bed> file of the merge isoforms.expressionfolder: contains all assembled <.gtf> with expression metrics computed by StringTie.ref_transcripts_annotation.bed: <.bed> file containing the coordinates of the genes specified by the user in the reference transcript file. Used for the bedtools intersect option to filter transcripts for these genes.ref_transcripts_annotation.gtf: <.gtf> annotation file filtered out of reference transcrits specified by the user from ensembl annot.SOSTAR_annotation_table_results.xlsx: final output file containing descriptive annotation and expression metrics of each transcript in the cohort (see SOSTAR annotation output file section for more informations).
The SOSTAR annotation module can be used as a stand-alone tool, as it is a simple Python script.
Using <.gtf> files of assembled isoforms from any assembly method, it provides
a descriptive and comprehensive annotation of each assembled isoform.
This module is compatible with any gtf file using the tag attribute gene_name that match
to the name of reference transcript feature provided by user.
python3 scripts/SOSTAR.py -I {INPUT} -O {OUTPUT} -R {REF_GTF_COORD}
Options:
-
-I,--input: /Path/to/gtf files -
-O,--output: /Path/to/output directory -
-R,--ref_gtf_coord: gtf annotation file filtered on reference transcripts used for the annotation
SOSTAR generates a spreadsheet file in <.xlsx> format:
| transcript_id | chr | start | end | strand | gene | annot_ref | annot_find | barcode01 | barcode13 | barcode25 | barcode37 | occurence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MSTRG.2.33 | 17 | 41188583 | 41277563 | - | BRCA1 | 1-24 | ▼1(176)-Δ11q(3309)-▼24(7729) | 29,96 | 9,56 | 5,87 | 10,25 | 4 |
| ENST00000491747.2 | 17 | 41197695 | 41277373 | - | BRCA1 | 1-24 | Δ1(14),Δ1q(6)-Δ11q(3309)-Δ14p(3)-Δ24(1383) | 16,24 | 2,35 | 2,12 | 0,17 | 4 |
transcript_id: transcript identifierchr: chromosome of transcriptstart: start position of the transcriptend: end position of the transcriptstrand: strand of the transcriptgene: gene associated to the transcriptannot_ref: exon range of the reference transcriptannot_find: SOSTAR annotation of transcript (see section SOSTAR nomenclature for more details).sample: transcript coverageoccurence: number of transcript occurrences in cohort
Isoforms are described relative to reference transcripts (provided by user) by an annotation including only the alternative splicing events. Some conventions were established to annotate the alternative splicing events:
| symbol | definition |
|---|---|
| ∆ | skipping of a reference exon |
| ▼ | inclusion of a reference intron |
| p | shift of an acceptor site |
| q | shift of a donnor site |
| (37) | number of skipped or retained nucleotides |
| [p23, q59] | relative positions of new splice sites |
| exo | exonization of an intronic sequence |
| int | intronization of an exonic sequence |
| - | continuous event |
| , | discontinuous event |
Nomenclature example:
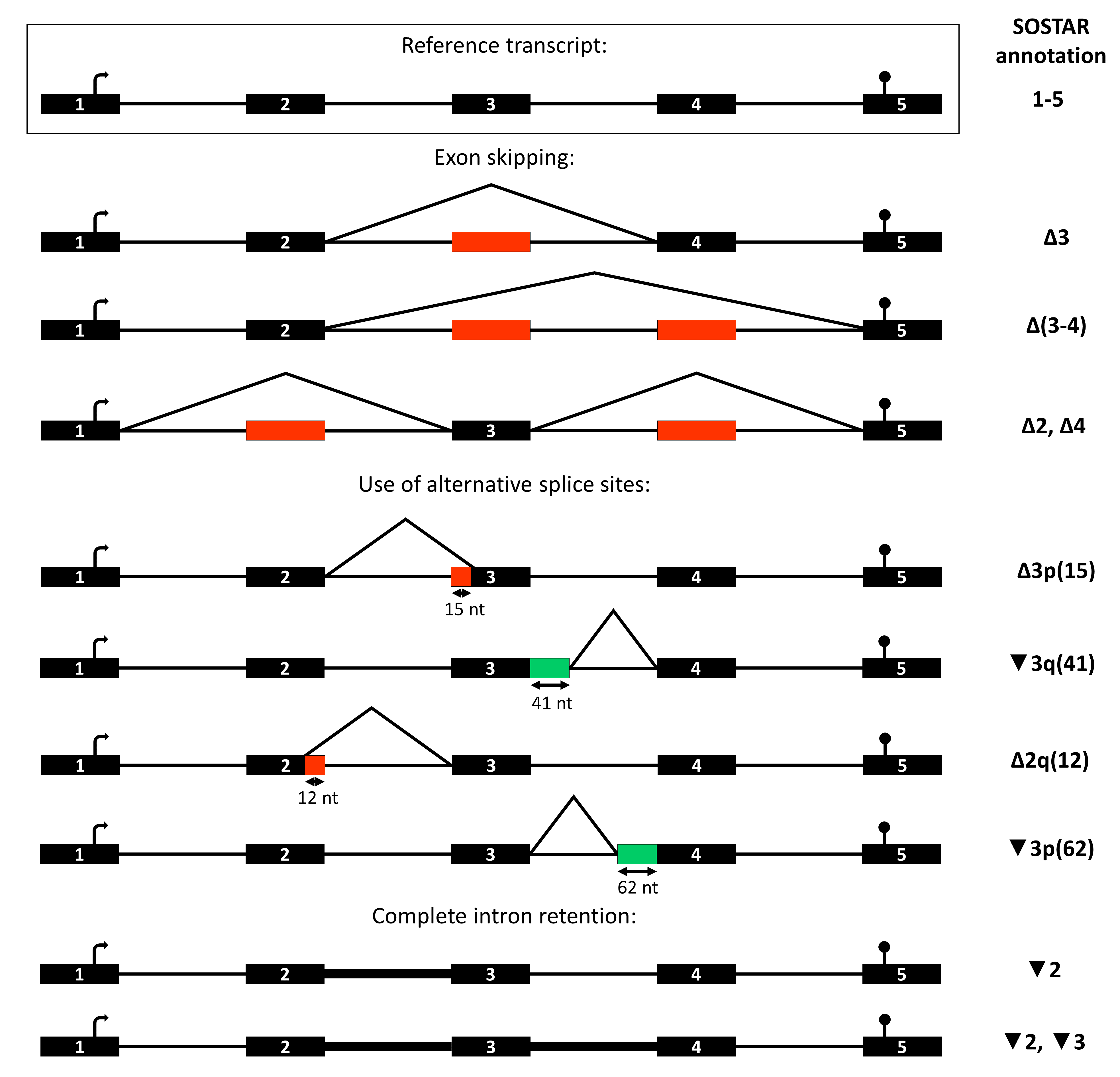 Black boxes: exon, black lines: intron, red boxes: exon (or part of exon) skipping, green boxes: novel exon (or part of exon).
Black boxes: exon, black lines: intron, red boxes: exon (or part of exon) skipping, green boxes: novel exon (or part of exon).
Camille AUCOUTURIER @AUCAM
This project is under GPL-3.0 License, see LICENCE for more details.