The goal of tidyphreeqc is to provide a more useful interface to the existing phreeqc package.
You can install tidyphreeqc from github with:
# install.packages("devtools")
devtools::install_github("paleolimbot/tidyphreeqc")Running PHREEQC is accomplished using the phr_run() function, which
calls the program and generates the output. The function accepts
character vectors of input, which can be generated using intput helper
functions such as phr_solution(), phr_selected_output(),
phr_equilibrium_phases(), and phr_reaction_temperature() (or roll
your own input using phr_input_section()).
library(tidyphreeqc)
phr_run(
phr_solution(pH = 7, temp = 25)
)
#> <phr_run_output>
#> PHREEQC run with 0 selected output(s)
#> Raw output at '/var/folders/bq/2rcjstv90nx1_wrt8d3gqw6m0000gn/T//RtmpbgYQSX/file1ac6bd49a06'To get the results as a data frame, we need to supply a
phr_selected_output() to the input file.
phr_run(
phr_solution(pH = 7, temp = 25),
phr_selected_output(pH = TRUE, temp = TRUE, activities = c("OH-", "H+", "O2"))
)
#> <phr_run_output>
#> PHREEQC run with 1 selected output(s)
#> Raw output at '/var/folders/bq/2rcjstv90nx1_wrt8d3gqw6m0000gn/T//RtmpbgYQSX/file1ac6709cbbca'
#> as_tibble():
#> # A tibble: 1 x 13
#> selected_output sim state soln dist_x time step pH pe
#> <chr> <int> <chr> <int> <dbl> <dbl> <int> <dbl> <dbl>
#> 1 n1 1 i_so… 1 NA NA NA 7 4
#> # … with 4 more variables: `temp(C)` <dbl>, `la_OH-` <dbl>, `la_H+` <dbl>,
#> # la_O2 <dbl>To find the distribution of a few solutions, you can generate a list of
solutions using phr_solution_list().
phr_run(
phr_solution_list(pH = 5:8, temp = 12:25),
phr_selected_output(pH = TRUE, temp = TRUE, activities = c("OH-", "H+", "O2"))
)
#> <phr_run_output>
#> PHREEQC run with 1 selected output(s)
#> Raw output at '/var/folders/bq/2rcjstv90nx1_wrt8d3gqw6m0000gn/T//RtmpbgYQSX/file1ac62fc49e88'
#> as_tibble():
#> # A tibble: 56 x 13
#> selected_output sim state soln dist_x time step pH pe
#> <chr> <int> <chr> <int> <dbl> <dbl> <int> <dbl> <dbl>
#> 1 n1 1 i_so… 1 NA NA NA 5 4
#> 2 n1 1 i_so… 2 NA NA NA 6 4
#> 3 n1 1 i_so… 3 NA NA NA 7 4
#> 4 n1 1 i_so… 4 NA NA NA 8 4
#> 5 n1 1 i_so… 5 NA NA NA 5 4
#> 6 n1 1 i_so… 6 NA NA NA 6 4
#> 7 n1 1 i_so… 7 NA NA NA 7 4
#> 8 n1 1 i_so… 8 NA NA NA 8 4
#> 9 n1 1 i_so… 9 NA NA NA 5 4
#> 10 n1 1 i_so… 10 NA NA NA 6 4
#> # … with 46 more rows, and 4 more variables: `temp(C)` <dbl>,
#> # `la_OH-` <dbl>, `la_H+` <dbl>, la_O2 <dbl>Some elements (for example, mercury) aren’t included in the base
database. There are a number of databases included in the PHREEQC
package, that you can choose by specifying the db argument of
phr_run(). One that includes mercury is the “minteq” database.
phr_run(
phr_solution(pH = 7, temp = 25, Hg = 0.1),
phr_selected_output(
activities = c("Hg", "Hg2+2", "Hg(OH)2", "Hg(OH)2", "HgOH+", "Hg(OH)3-")
),
db = "minteq"
)
#> <phr_run_output>
#> PHREEQC run with 1 selected output(s)
#> Raw output at '/var/folders/bq/2rcjstv90nx1_wrt8d3gqw6m0000gn/T//RtmpbgYQSX/file1ac6ffc9491'
#> as_tibble():
#> # A tibble: 1 x 14
#> selected_output sim state soln dist_x time step pH pe la_Hg
#> <chr> <int> <chr> <int> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
#> 1 n1 1 i_so… 1 NA NA NA 7 4 -4.00
#> # … with 4 more variables: `la_Hg2+2` <dbl>, `la_Hg(OH)2` <dbl>,
#> # `la_HgOH+` <dbl>, `la_Hg(OH)3-` <dbl>result <- phr_run(
phr_solution_list(pH = seq(0, 14, 0.5), pe = seq(-14, 22, 0.5), Hg = 0.1),
phr_selected_output(
activities = c("Hg", "Hg2+2", "Hg(OH)2", "Hg(OH)2", "HgOH+", "Hg(OH)3-")
),
db = "minteq"
)
result
#> <phr_run_output>
#> PHREEQC run with 1 selected output(s)
#> Raw output at '/var/folders/bq/2rcjstv90nx1_wrt8d3gqw6m0000gn/T//RtmpbgYQSX/file1ac69c10ab8'
#> as_tibble():
#> # A tibble: 1,189 x 14
#> selected_output sim state soln dist_x time step pH pe la_Hg
#> <chr> <int> <chr> <int> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
#> 1 n1 1 i_so… 1 NA NA NA 14 -13.5 -3.92
#> 2 n1 1 i_so… 2 NA NA NA 13.5 -13 -3.98
#> 3 n1 1 i_so… 3 NA NA NA 14 -13 -3.92
#> 4 n1 1 i_so… 4 NA NA NA 13 -12.5 -3.99
#> 5 n1 1 i_so… 5 NA NA NA 13.5 -12.5 -3.98
#> 6 n1 1 i_so… 6 NA NA NA 14 -12.5 -3.92
#> 7 n1 1 i_so… 7 NA NA NA 12.5 -12 -4.00
#> 8 n1 1 i_so… 8 NA NA NA 13 -12 -3.99
#> 9 n1 1 i_so… 9 NA NA NA 13.5 -12 -3.98
#> 10 n1 1 i_so… 10 NA NA NA 14 -12 -3.92
#> # … with 1,179 more rows, and 4 more variables: `la_Hg2+2` <dbl>,
#> # `la_Hg(OH)2` <dbl>, `la_HgOH+` <dbl>, `la_Hg(OH)3-` <dbl>library(tidyverse)
#> Registered S3 method overwritten by 'rvest':
#> method from
#> read_xml.response xml2
#> ── Attaching packages ───────────────────────────────────────────────────────────────────── tidyverse 1.2.1 ──
#> ✔ ggplot2 3.2.1 ✔ purrr 0.3.2
#> ✔ tibble 2.1.3 ✔ dplyr 0.8.1
#> ✔ tidyr 0.8.3 ✔ stringr 1.4.0
#> ✔ readr 1.3.1 ✔ forcats 0.4.0
#> ── Conflicts ──────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
result_long <- result %>%
as_tibble() %>%
gather(key = "species", value = "log_activity", starts_with("la_")) %>%
mutate(species = str_remove(species, "^la_"))
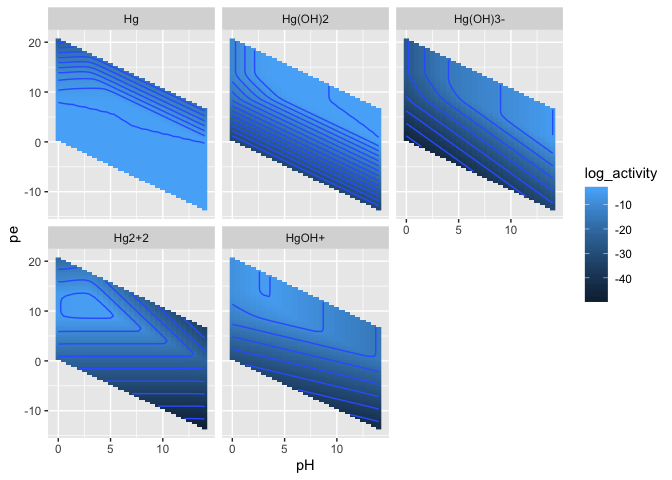
result_long %>%
ggplot(aes(x = pH, y = pe, fill = log_activity)) +
geom_raster() +
stat_contour(aes(z = log_activity)) +
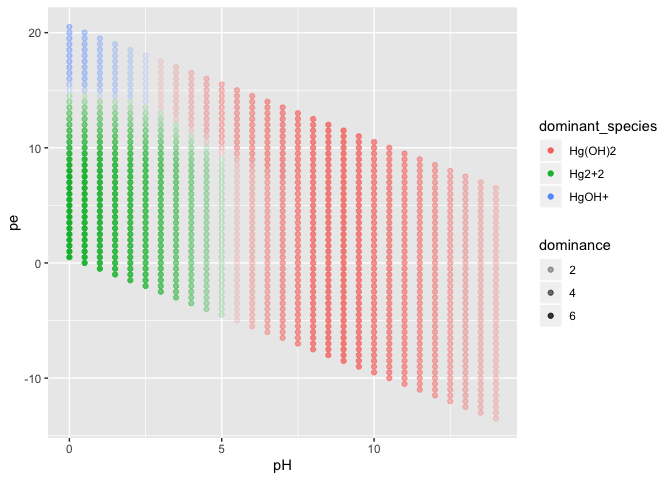
facet_wrap(~species)result_long %>%
filter(species != "Hg") %>%
group_by(pH, pe) %>%
summarise(
dominant_species = species[which.max(log_activity)],
dominant_log_act = max(log_activity),
dominance = max(log_activity) - max(setdiff(log_activity, max(log_activity)))
) %>%
ggplot(aes(pH, pe, col = dominant_species, alpha = dominance)) +
geom_point()