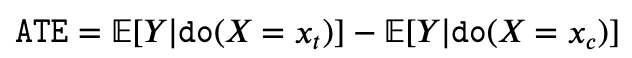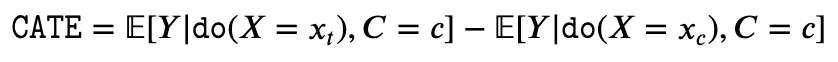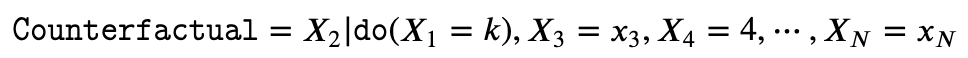🔥🔥Updates🔥🔥
- Added GES, LINGAM, GIN for tabular data causal discovery
- Added markov blanket discovery algorithm (grow-shrink) for tabular data
- Added support for heterogeneous data (mixed discrete and continuous variables)
- Added benchmarking modules for tabular and time series data, that will help the research community benchmark new and existing causal discovery algorithms against various challenges in the data (graph sparsity, sample complexity, variable complexity, SNR, noise type, etc)
- Root cause analysis for tabular and time series data
- Introduction
- Comparison with Related Libraries
- Causal Discovery
- Causal Inference
- Installation
- Quick Tutorial
- User Inferface
- Documentation
- Technical Report and Citing Salesforce CausalAI
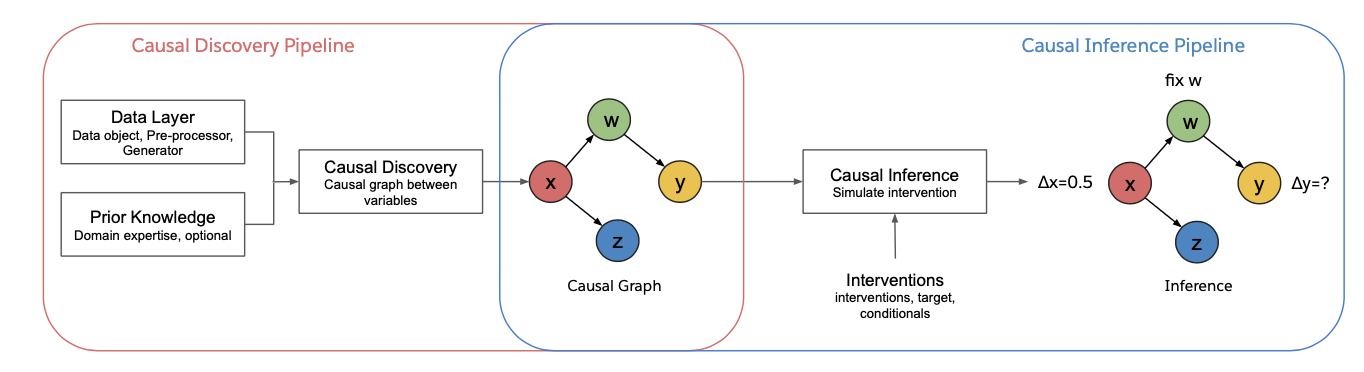
Salesforce CausalAI is an open-source Python library for causal analysis using observational data. It supports causal discovery and causal inference for tabular and time series data (see figure above), of discrete, continuous and heterogeneous data types. It also supports Markov Blanket discovery algorithms. This library includes algorithms that handle linear and non-linear causal relationship between variables, and uses multi-processing for speed-up. We also include a data generator capable of generating synthetic data with specified structural equation model for both the aforementioned data formats and types, that helps users control the ground-truth causal process while investigating various algorithms. CausalAI includes benchmarking modules for tabular and time series data, that users can use to compare different causal discovery algorithms, as well as evaluate the performance of a particular algorithm across datasets with different challenges. Specifically, users can evaluate the performance of causal discovery algorithms on synthetic data with varying graph sparsity, sample complexity, variable complexity, SNR, noise type, and max lag (time series data). Finally, we provide a user interface (UI) that allows users to perform causal analysis on data without coding. The goal of this library is to provide a fast and flexible solution for a variety of problems in the domain of causality.
Some of the key features of CausalAI are:
- Algorithms: Support for causal discovery, causal inference, Markov blanket discovery, and Root Cause Analysis (RCA).
- Data: Causal analysis on tabular and time series data, of discrete, continuous and heterogeneous types.
- Missing Values: Support for handling missing/NaN values in data.
- Data Generator: A synthetic data generator that uses a specified structural equation model (SEM) for generating tabular and time series data. This can be used for evaluating and comparing different causal discovery algorithms since the ground truth values are known.
- Distributed Computing: Use of multi-processing using the Python Ray library, that can be optionally turned on by the user when dealing with large datasets or number of variables for faster compute.
- Targeted Causal Discovery: In certain cases, we support targeted causal discovery, in which the user is only interested in discovering the causal parents of a specific variable of interest instead of the entire causal graph. This option reduces computational overhead.
- Visualization: Visualize tabular and time series causal graphs.
- Prior Knowledge: Incorporate any user provided partial prior knowledge about the causal graph in the causal discovery process.
- Benchmarking: Benchmarking module for comparing different causal discovery algorithms, as well as evaluating the performance of a particular algorithm across datasets with different challenges.
- Code-free UI: Provide a code-free user interface in which users may directly upload their data and perform their desired choice of causal analysis algorithm at the click of a button.
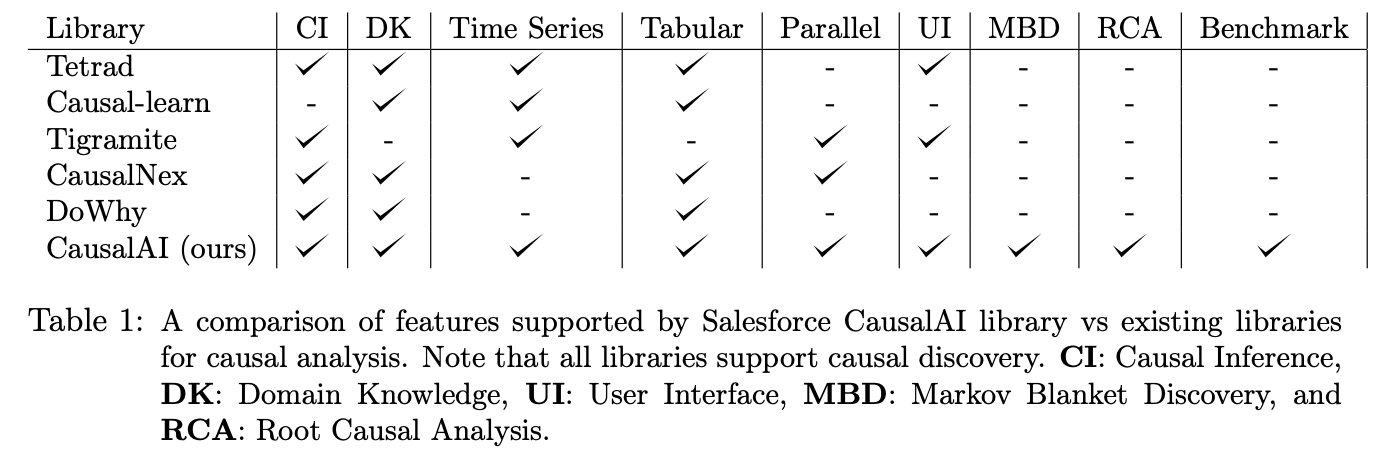
The table below provides a visual overview of how CausalAI's key features compare to other libraries for causal analysis.
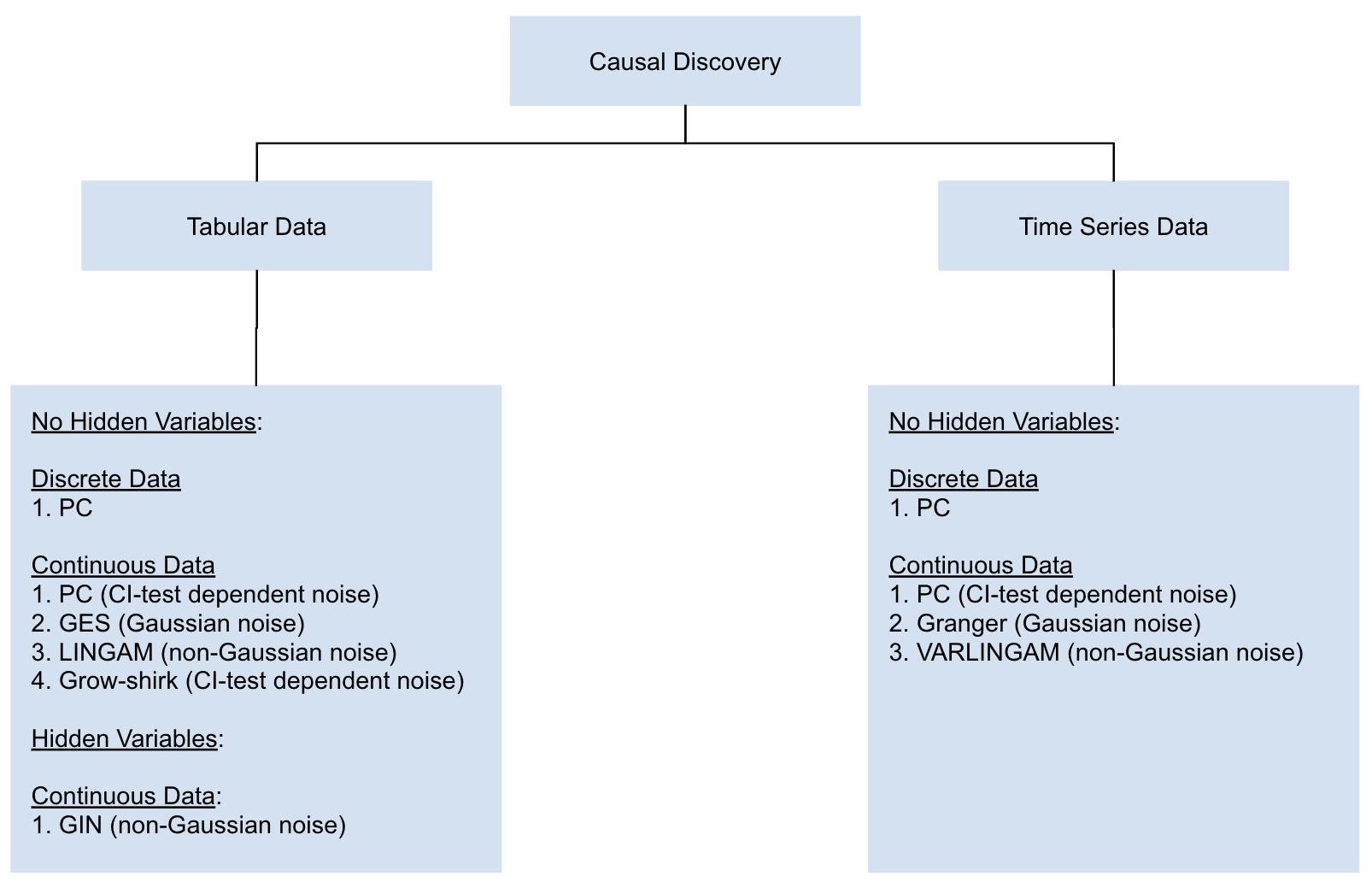
We support the following causal discovery algorithm categorized by their assumptions on whether hidden variables are allowed, whether the data is discrete or contiuous, and the type of noise in the data.
For continuous data, PC algorithm and Grow-shrink support both linear and non-linear causal relationships (depending on the CI test used). All other algorithms support linear relationships.
We support the following causal inference estimations for tabular and time series data of continuous and discrete types:
- Average Treatment Effect (ATE): ATE aims to determine the relative expected difference in the value of Y when we intervene X to be x_t compared to when we intervene X to be x_c. Here x_t and x_c are respectively the treatment value and control value.
- Conditional Average Treatment Effect (CATE): CATE is similar to ATE, except that in addition to intervetion on X, we also condition on some set of variables C taking value c. Notice here that X is intervened but C is not.
- Counterfactual: Counterfactuals aim at estimating the effect of an intervention on a specific instance or sample. Suppose we have a specific instance of a system of random variables (X_1, X_2,...,X_N) given by (X_1=x_1, X_2=x_2,...,X_N=x_N), then in a counterfactual, we want to know the effect an intervention (say) X_1=k would have had on some other variable(s) (say X_2), holding all the remaining variables fixed.
Depending on whether the relationship between variables is linear or non-linear, the user may specify a linear or non-linear prediction model respectively in the inference module.
Prior to installing the library, create a conda environment with Python 3.9 or a later version. This can be done by executing conda create -n causal_ai_env python=3.9. Activate this environment by executing conda activate causal_ai_env. To install Salesforce CausalAI, git clone the library, go to the root directory of the repository, and execute pip install ..
Before importing and calling the library, or launching the UI, remember to first activate the conda environemnt.
Let's suppose we have some observational tabular data, and we want to perform causal discovery using the PC algorithm. The following code illustrates this using data that is synthetically generated using the CausalAI library:
# Causal Discovery using PC algorithm on Tabular Data
from causalai.models.tabular.pc import PCSingle, PC
from causalai.models.common.CI_tests.partial_correlation import PartialCorrelation
from causalai.data.data_generator import DataGenerator # for generating data randomly
from causalai.models.common.prior_knowledge import PriorKnowledge
from causalai.data.tabular import TabularData # tabular data object
from causalai.data.transforms.time_series import StandardizeTransform
#### Generate a ground truth causal graph and data radom using it, for illustration purposes
fn = lambda x:x # non-linearity
coef = 0.1
# Structural equation model (SEM) defining the ground truth causal graph
sem = {
'a': [],
'b': [('a', coef, fn), ('f', coef, fn)], # b = coef* fn(a) + coef* fn(f) + noise
'c': [('b', coef, fn), ('f', coef, fn)],
'd': [('b', coef, fn), ('g', coef, fn)],
'e': [('f', coef, fn)],
'f': [],
'g': [],
}
T = 5000 # number of samples
data_array, var_names, graph_gt = DataGenerator(sem, T=T, seed=0, discrete=False)
# data_array is a (T x 7) NumPy array
# var_names = ['a', 'b', 'c', 'd', 'e', 'f', 'g']
# graph_gt is a Python dictionary
### standardize data and create a CausalAI Tabular data object
StandardizeTransform_ = StandardizeTransform()
StandardizeTransform_.fit(data_array)
data_trans = StandardizeTransform_.transform(data_array)
data_obj = TabularData(data_trans, var_names=var_names)
### Run PC algorithm
# provide optional (use None) prior knowledge saying b->a is forbidden.
prior_knowledge = PriorKnowledge(forbidden_links={'a': ['b']})
pvalue_thres = 0.01
CI_test = PartialCorrelation()
pc = PC(
data=data_obj,
prior_knowledge=prior_knowledge,
CI_test=CI_test,
use_multiprocessing=False
)
result = pc.run(pvalue_thres=pvalue_thres, max_condition_set_size=2)
# print estimated causal graph
graph_est={n:[] for n in result.keys()}
for key in result.keys():
parents = result[key]['parents']
graph_est[key].extend(parents)
print(f'{key}: {parents}')
########### prints
# a: []
# b: ['d', 'a', 'c', 'f']
# c: ['f', 'b']
# d: ['g', 'b']
# e: ['f']
# f: ['e', 'b', 'c']
# g: ['d']
###########
### Evaluate the estimated causal graph given we have ground truth in this case
from causalai.misc.misc import plot_graph, get_precision_recall
precision, recall, f1_score = get_precision_recall(graph_est, graph_gt)
print(f'Precision {precision:.2f}, Recall: {recall:.2f}, F1 score: {f1_score:.2f}')
# Precision 0.64, Recall: 1.00, F1 score: 0.67Now let's suppose we have some observational tabular data and the causal graph (not the SEM) for this data, and we want to estimate causal inference effects (specifically ATE in this example) on a desired target variable given treatment variables. The following code illustrates this using data that is synthetically generated using the CausalAI library:
# Causal Inference on Tabular Data using Backdoor method and in-house CausalPath method
from causalai.data.data_generator import DataGenerator
from causalai.models.tabular.causal_inference import CausalInference
from sklearn.linear_model import LinearRegression
import numpy as np
def define_treatments(name, t,c):
treatment = dict(var_name=name,
treatment_value=t,
control_value=c)
return treatment
#### Generate a ground truth causal graph and data radom using it, for illustration purposes
fn = lambda x:x # non-linearity
coef = 0.5
# Structural equation model (SEM) defining the ground truth causal graph
sem = {
'a': [],
'b': [('a', coef, fn), ('f', coef, fn)],
'c': [('b', coef, fn), ('f', coef, fn)],
'd': [('b', coef, fn), ('g', coef, fn)],
'e': [('f', coef, fn)],
'f': [],
'g': [],
}
T = 5000 # number of samples
data, var_names, graph_gt = DataGenerator(sem, T=T, seed=0, discrete=False)
# data_array is a (T x 7) NumPy array
# var_names = ['a', 'b', 'c', 'd', 'e', 'f', 'g']
# graph_gt is a Python dictionary
### Define treatment variables, and their treatment and control values
t1='a' # treatment variable
t2='b' # treatment variable
target = 'c' # target variable; Notice c does not depend on a if we intervene on b.
target_var = var_names.index(target)
# treatment values
intervention11 = 100*np.ones(T)
intervention21 = 10*np.ones(T)
# control values
intervention12 = -0.*np.ones(T)
intervention22 = -2.*np.ones(T)
treatments = [define_treatments(t1, intervention11,intervention12),\
define_treatments(t2, intervention21,intervention22)]
### Perform Causal Inference
CausalInference_ = CausalInference(data, var_names, graph_gt, LinearRegression , discrete=False, method='causal_path')
ate, y_treat,y_control = CausalInference_.ate(target, treatments)
print(f'Estimated ATE using causal_path method: {ate:.2f}')
# Estimated ATE using causal_path method: 6.38
CausalInference_ = CausalInference(data, var_names, graph_gt, LinearRegression , discrete=False, method='backdoor')
ate, y_treat,y_control = CausalInference_.ate(target, treatments)
print(f'Estimated ATE using backdoor method: {ate:.2f}')
# Estimated ATE using backdoor method: 7.21
### Compute the True ATE given we can generate data with intervention in this case since it is synthetically generated
intervention_data1,_,_ = DataGenerator(sem, T=T, seed=0,
intervention={t1:intervention11, t2:intervention21})
intervention_data2,_,_ = DataGenerator(sem, T=T, seed=0,
intervention={t1:intervention12, t2:intervention22})
true_effect = (intervention_data1[:,target_var] - intervention_data2[:,target_var]).mean()
print("True ATE = %.2f" %true_effect)
# True ATE = 6.00For more tutorials on other supported functionalities of CausalAI, please see
tutorials.
We provide a UI for users to directly upload their data and run causal discovery and causal inference algorithms without the need to write any code. An introduction to the UI can be found here.
In order to launch the UI, go to the root directory of the library and execute ./launch_ui.sh, and open the url specified in the terminal in a browser. In order to terminate the UI, press Ctrl+c in the terminal where the UI was launched, and then execute ./exit_ui.sh.
For Jupyter notebooks with exmaples, see
tutorials. Detailed API documentation with tutorials can be found here. The
technical report describes the implementation details of the algorithms along with their assumptions and also covers important aspects of the API. Further, it also presents experimental results that demosntrate the speed and performance our library compared with some of the existing libraries.
You can find more details in our technical report
If you're using Salesforce CausalAI in your research or applications, please cite using this BibTeX:
@article{salesforce_causalai23,
title={Salesforce CausalAI Library: A Fast and Scalable framework for Causal Analysis of Time Series and Tabular Data},
author={Arpit, Devansh and Fernandez, Matthew, and Feigenbaum, Itai and Yao, Weiran and Liu, Chenghao and Yang, Wenzhuo and Josel, Paul and Heinecke, Shelby and Hu, Eric and Wang, Huan and Hoi, Stephen and Xiong, Caiming and Zhang, Kun and Niebles, Juan Carlos},
year={2023},
eprint={arXiv preprint arXiv:2301.10859},
archivePrefix={arXiv},
primaryClass={cs.LG}
}