A Snakemake-wrapper for evaluating de novo bacterial isolate genome assemblies, e.g. from Oxford Nanopore (ONT) or Illumina sequencing, using multiple programs. The results are summarized in a HTML report.
The workflow is published in Snakemake workflows for long-read bacterial genome assembly and evaluation in GigaByte.
Following programs are included in the workflow:
- pomoxis assess_assembly and assess_homopolymers
- dnadiff from the mummer package
- NucDiff
- QUAST
- BUSCO
- ideel, which uses prodigal and diamond
- bakta
Clone repository, for example:
git clone https://github.com/pmenzel/score-assemblies.git /opt/software/score-assemblies
Create a new conda environment containing all necessary programs:
conda env create -n score-assemblies --file /opt/software/score-assemblies/env/environment.yaml
and activate the environment:
conda activate score-assemblies
First, prepare a data folder, which must contain subfolders assemblies/ containing the
assemblies.
Additionally, the sub-foldersreferences/ and references-protein/ can contain reference genomes and reference proteins with which the assemblies and predicted proteins will be compared.
For example:
.
├── assemblies
│ ├── example-mtb_flyehq4.fa
│ ├── example-mtb_flyehq4+medaka.fa
│ ├── example-mtb_flyehq.fa
│ ├── example-mtb_flyehq+racon4.fa
│ ├── example-mtb_flyehq+racon4+medaka.fa
│ ├── example-mtb_raven4.fa
│ ├── example-mtb_raven4+medaka.fa
│ ├── example-mtb_raven4+medaka+pilon.fa
│ └── example-mtb_unicycler.fa
├── references
│ └── AL123456.3.fa
└── references-protein
└── AL123456.3.faa
NB: The assembly and reference FASTA files need to have the .fa extension and protein reference FASTA files need to have the extension .faa.
This is the same folder structure used by ont-assembly-snake, i.e. score-assemblies can be run directly in the same folder.
To run the workflow, e.g. with 20 threads, use this command:
snakemake -s /opt/software/score-assemblies/Snakefile --cores 20 --use-conda
Output files of each program will be written to various folders in score-assemblies-data/.
If no references are supplied, then only ideel and BUSCO are done, otherwise score-assemblies will run these programs on each assembly:
Each assembly will be compared against each reference genome using the
assess_assembly and assess_homopolymers scripts from
pomoxis. Additionally to the tables
and plots generated by these programs, summary plots for each reference genome will be plotted
in score-assemblies-data/pomoxis/<reference>_assess_assembly_all_meanQ.pdf.
Set the lineage via the snakemake call:
snakemake -s /opt/software/score-assemblies/Snakefile --cores 20 --config busco_lineage=bacillales
If not set, the default lineage bacteria will be used.
Available datasets can be listed with busco --list-datasets
The number of complete, fragmented and missing BUSCOs per assembly is tabulated in the file score-assemblies-data/busco/all_stats.tsv and also drawn as dotplot in score-assemblies-data/busco/busco_stats.pdf.
Each assembly is compared with each reference and the output files will be
located in score-assemblies-data/dnadiff/<reference>/<assembly>-dnadiff.report. The values for
AvgIdentity (from 1-to-1 alignments) and TotalIndels are extracted from these files and are plotted
for each reference in score-assemblies-data/dnadiff/<reference>_dnadiff_stats.pdf.
Each assembly is compared with each reference and the output files will be
located in the folder score-assemblies-data/nucdiff/<reference>/<assembly>-nucdiff/. The values for
Insertions, Deletions, and Substitutions are extracted from the file results/nucdiff_stat.out and are drawn
for each reference in score-assemblies-data/nucdiff/<reference>_nucdiff_stats.pdf.
One QUAST report is generated for each reference genome, containing the results for all assemblies.
The report files are located in score-assemblies-data/quast/<reference>/report.html.
The main report file score-assemblies-report.html also links the these individual reports.
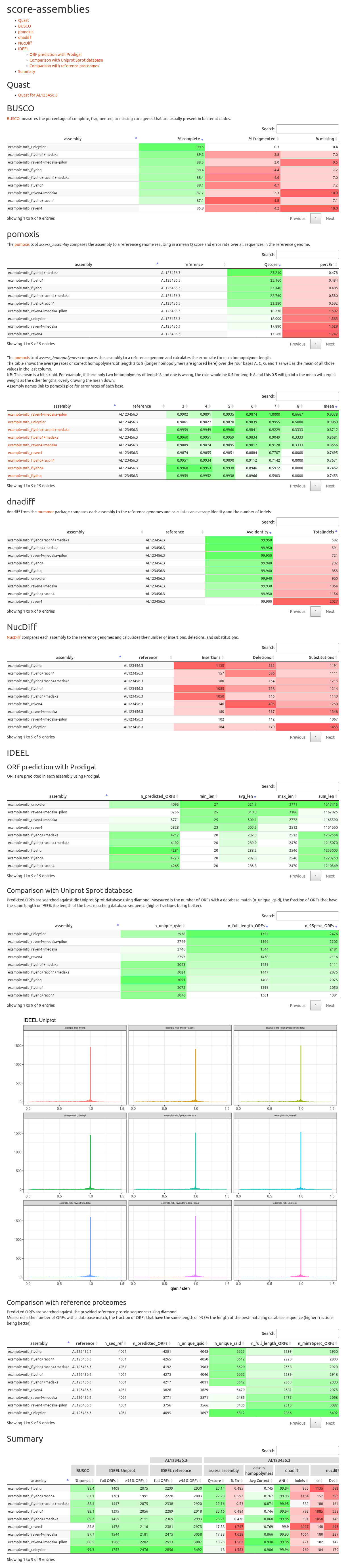
Open reading frames are predicted from each assembly via Prodigal and are
search in the Uniprot sprot database with diamond, retaining the best alignment
for each ORF. For each assembly, the distribution of the ratios between length
of the ORF and the matching database sequence are plotted to ideel/ideel_uniprot_histograms.pdf and ideel/ideel_uniprot_boxplots.pdf.
Additionally, diamond alignments are done between the predicted ORFs and the supplied reference proteins and ratios are plotted to
score-assemblies-data/ideel/<reference>_ideel_histograms.pdf and score-assemblies-data/ideel/<reference>_ideel_boxplots.pdf.
bakta is only run when specified as extra config argument in the snakemake call:
snakemake -s /opt/software/score-assemblies/Snakefile --cores 20 --use-conda --config bakta=1
The bakta outfiles files are written to in the folder score-assemblies-data/bakta/<assembly>/.
NB: It takes a long time to download the bakta database and run bakta on all assemblies.
All measurements are summarized in a HTML page in score-assemblies-report.html.