A Python project to download, process and visualize event-based seismic waveform segments, specifically designed to manage big volumes of data.
The key aspects with respect to widely-used similar applications is the use of a Relational database management system (RDBMS) to store downloaded data and metadata. Note the program supports SQLite and Postgres RDBMS. For massive downloads (as a rule of thumb: ≥ 1 million segments) we suggest to use Postgres, and we strongly suggest to run the program on computers with at least 16GB of RAM.
The main advantages of this approach are:
-
Storage efficiency: no huge amount of files, no complex, virtually unusable directory structures. Moreover, a database prevents data and metatada inconsistency by design, and allows more easily to track what has already been downloaded in order to customize and improve further downloads
-
Simple Python objects representing stored data and relationships, easy to work with in any kind of custom code. For instance, a segment is represented by a
Segmentobject with its data, metadata and related objects easily accessible through its attributes, e.g.,segment.stream(),segment.maxgap_numsamples,segment.event.magnitude,segment.station.network,segment.channel.orientation_codeand so on. -
A powerful segments selection made even easier by means of a simplified syntax: map any attribute described above to a selection expression (e.g.
segment.event.magnitude: "[4, 5)") and with few lines you can compose complex database queries such as e.g., "get all downloaded segments within a given magnitude range, with no gaps, enough data (related to the requested data), from broadband channels only and a given specific network"
Each download is highly customizable with several parameters for any step required (event, data center, station and waveform data download). In addition, as data is never downloaded per se, Stream2segment helps the whole workflow with:
-
An integrated processing environment to get any user-dependent output (e.g., tabular output such as CSV or HDF files). Write your segment selection in a configuration (YAML) file, and your own code in a processing (Python) module. Pass both files to the
s2s processcommand, and Stream2segment takes care of executing the code on all selected segments, interacting with the database for you while displaying progress bars, estimated available time, and handling errors. -
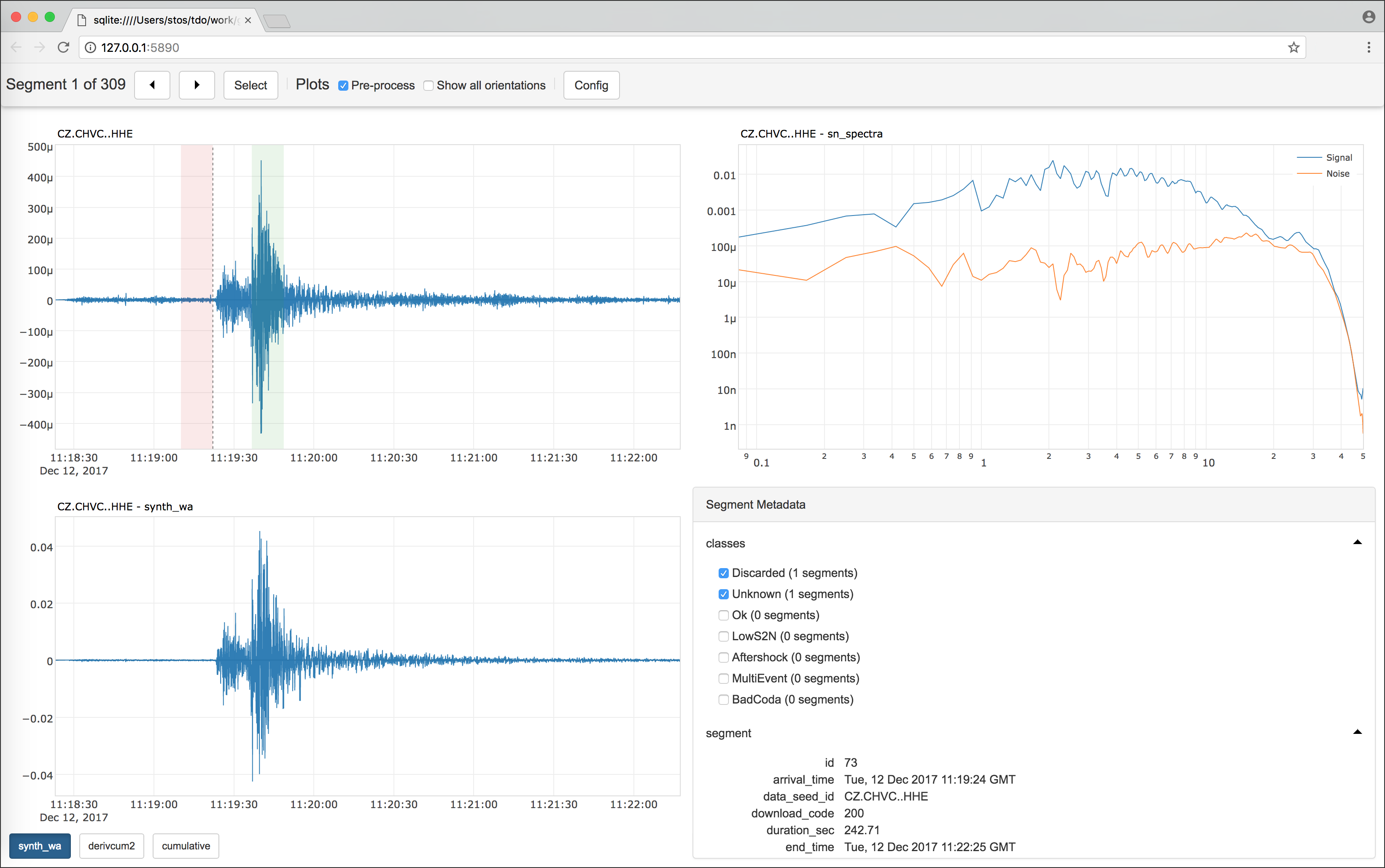
A visualization tool to show any kind of user defined plot from each selected segment. Similar to the processing case above, write your selection in the configuration file and the code of your own plots in the processing (or any Python) module. Pass both files to the
s2s showcommand, and Stream2segment takes care of visualizing your plots in a web browser Graphical User Interface (GUI) with no external programs required. The GUI can also be used to label segments with user-defined classes in order to refine the segments selection later, or for creating datasets in machine-learning supervised classification problems -
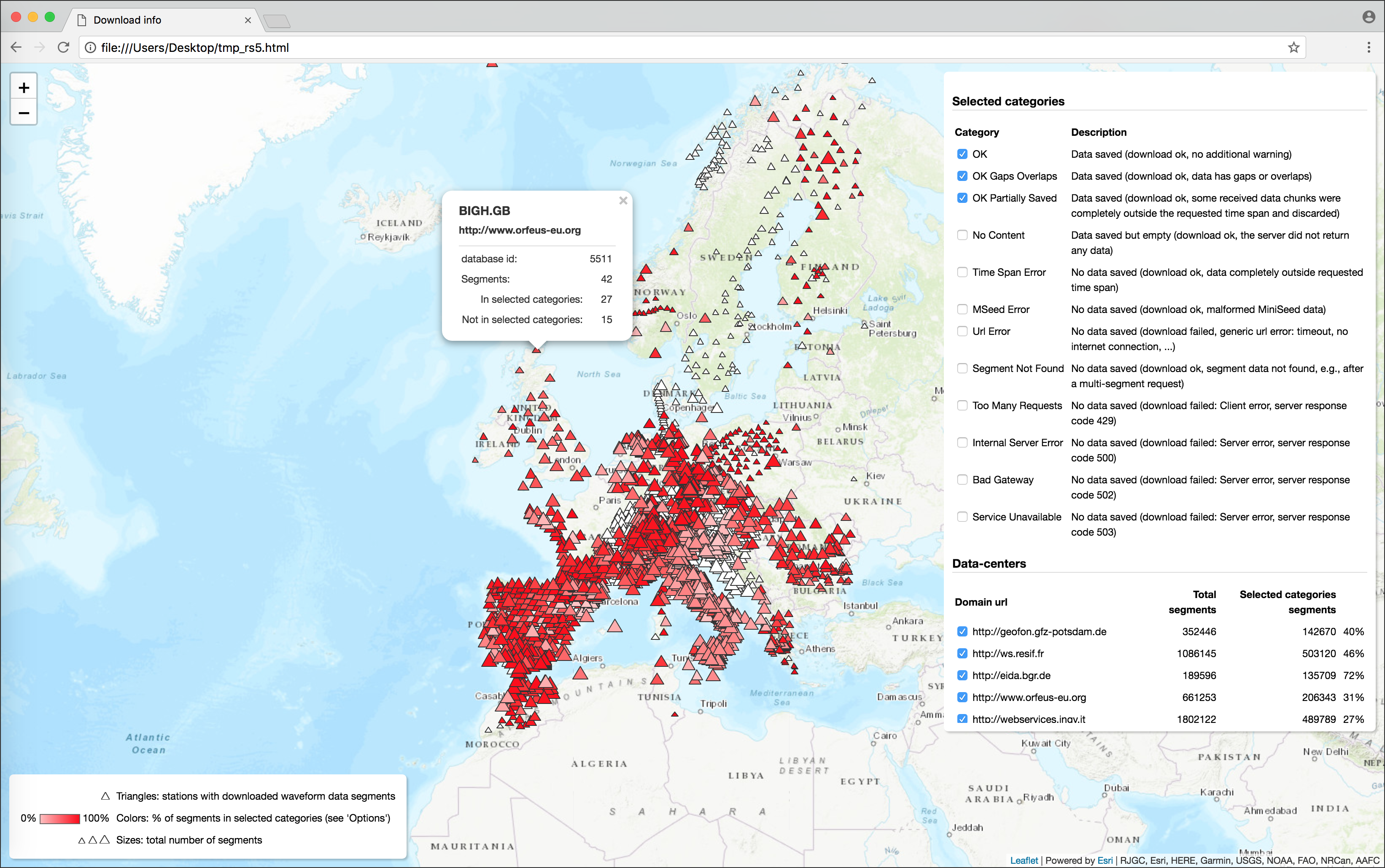
Several utilities to interact with the database, print download reports or show interactive stats on web GUI maps (see command
s2s utils)The GUI produced with the showcommandThe dynamic HTML page produced with the utils dstatscommand(image linked from https://geofon.gfz-potsdam.de/software/stream2segment/) (image linked from https://geofon.gfz-potsdam.de/software/stream2segment/)
Citation (Software):
Zaccarelli, Riccardo (2018): Stream2segment: a tool to download, process and visualize event-based seismic waveform data. V. 2.7.3. GFZ Data Services.
http://doi.org/10.5880/GFZ.2.4.2019.002
Citation (Research article):
Riccardo Zaccarelli, Dino Bindi, Angelo Strollo, Javier Quinteros and Fabrice Cotton. Stream2segment: An Open‐Source Tool for Downloading, Processing, and Visualizing Massive Event‐Based Seismic Waveform Datasets. Seismological Research Letters (2019)
https://doi.org/10.1785/0220180314
A detailed documentation is available online in the
github wiki page, but
you can also simply start the program via the command init (
s2s init --help for details) which creates several examples files to run
the program. These files contain roughly the same online documentation
in form of comments to code and parameters that you can immediately start to
configure and modify. These files are:
- A download configuration file (in YAML syntax with all parameters documented)
to start the download routine:
s2s download -c <config_file> ...
- Two modules (Python files) with relative configurations (YAML files)
to be passed to the processing routine:
or the visualization routine:
s2s process -c <config_file> -p <processing_module> ...
s2s show -c <config_file> -p <processing_module> ...
- A Jupyter notebook tutorial with examples, for user who prefer this approach when working with downloaded data
This program has been installed and tested on Ubuntu (14 and later) and macOS (El Capitan and later).
In case of installation problems, we suggest you to proceed in this order:
- Look at Installation Notes to check if the problem has already ben observed and a solution proposed
- Google for the solution (as always)
- Ask for help
In this section we assume that you already have Python (3.5 or later) and the required database software. The latter should not needed if you use SQLite or if the database is already installed remotely, so basically you are concerned only if you need to download data locally (on your computer) on a Postgres database.
On MacOS (El Capitan and later) all required software is generally already preinstalled. We suggest you to go to the next step and look at the Installation Notes in case of problems (to install software on MacOS, we recommend to use brew).
Details
In few cases, on some computers we needed to run one or more of the following commands (it's up to you to run them now or later, only those really needed):
xcode-select --install
brew install openssl
brew install c-blosc
brew install git
Ubuntu does not generally have all required packages pre-installed. The bare minimum
of the necessary packages can be installed with the apt-get command:
sudo apt-get install git python3-pip python3-dev # python 3
Details
In few cases, on some computers we needed to run one or more of the following commands (it's up to you to run them now or later, only those really needed):
Upgrade gcc first:
sudo apt-get update
sudo apt-get upgrade gcc
Then:
sudo apt-get update
sudo apt-get install libpng-dev libfreetype6-dev \
build-essential gfortran libatlas-base-dev libxml2-dev libxslt-dev python-tk
Git-clone (basically: download) this repository to a specific folder of your choice:
git clone https://github.com/rizac/stream2segment.git
and move into package folder:
cd stream2segment
We strongly recommend to use Python virtual environment, because by isolating all Python packages we are about to install, we won't create conflicts with already installed packages.
Python (from version 3.3) has a built-in support for virtual environments - venv
(On Ubuntu, you might need to install it first
via sudo apt-get install python3-venv).
Make virtual environment in an stream2segment/env directory (env is a convention, but it's ignored by git commits so better keeping it. You can also use ".env" which makes it usually hidden in Ubuntu).
python3 -m venv ./env
To activate your virtual environment, type:
source env/bin/activate
or source env/bin/activate.csh (depending on your shell)
Activation needs to be done each time we will run the program. To check you are in the right env, type:
which pipand you should see it's pointing inside the env folder
Installation and activation with Anaconda (click to expand)
disclaimer: the lines below might be outdated. Please refer to the Conda documentation for details
Create a virtual environment for your project
- In the terminal client enter the following where yourenvname (like « env ») is the
name you want to call your environment, and replace x.x with the Python version you
wish to use. (To see a list of available Python versions first, type conda search
"^python$" and press enter.)
Conda create –n yourenvname python=x.x anaconda - Press « y » to proceed
Activate your virtual environment
$source activate env- To deactivate this environment, use
$source deactivate
Important reminders before installing:
- From now on you are supposed to be in the stream2segment directory, (where you cloned the repository) with your Python virtualenv activated
- In case of errors, check the Installation notes below
To install the package, you should run as usual pip install, but because
some required packages must unfortunately be installed in a specific order,
we implemented a script that handles that and can be invoked exactly as pip install:
./pipinstall <options...> .
script details (if you want to run each `pip install` command separately to have more control)
pipinstall is simply a shorthand for several pip install commands, run in these
specific order:
-
Install pre-requisites as
pip install --upgrade pip setuptools wheel -
Install numpy first (this is an obspy requirement): either
pip install numpyor, if you want to use a requirements file, extracting (e.g. viagrep) the specific numpy version in the file (e.g.numpy==1.15.4), and then executingpip install numpy==1.15.4 -
Running
pip installwith exactly the same arguments provided to the script. E.g.pipinstall -e .executespip install -e .
The pipinstall argument . means "install this directory" (i.e., stream2segment) and
can be enhanced with extra packages. For instance, if you want to install Jupyter in order
to work with Stream2segment downloaded data in a notebook, then type:
./pipinstall <options...> ".[jupyter]"
If you want to install additional packages needed for testing (install in dev mode) and be able to push code and/or run tests, then type:
./pipinstall <options...> ".[dev]"
(You can also provide both: ".[dev,jupyter]". Quotes were necessary on some
specific macOS with zsh, in other OSs or shell languages might not be needed)
The <options...> are the usual pip install options. The two more important are usually:
-e This makes the package editable.
A typical advantage of an editable package is that when you run git pull to fetch a new
version that does not need new requirements (e.g. a bugfix), you don't need to reinstall
it but the new version will be already available for use
-r ./requirements.txt: install requirements with specific versions. pip install by
default skips already installed requirements if they satisfy Stream2segment minimum
versions. With the -r option instead, requirements are installed with "tested" versions
(i.e., those "freezed" after successfully running tests), which should generally be safer
for obvious reasons. However, some versions in the requirements might not be (yet?) supported
in your computer, some might be in conflicts with the requirements of other packages you
installed in the virtualenv, if any. You can try this option and then remove it, in case
of problems. In any case, do not use this option if you plan to install other
stuff alongside stream2segemtn on the virtualenv.
(There is also a ./requirements.dev.txt
that installs also the dev-related packages, similar to ".[dev]", but with specific
exact versions.
The program is now installed. To double check the program functionalities, we suggest to run tests (see below) and report the problem in case of failure. In any case, before reporting a problem remember to check first the Installation Notes
Stream2segment has been highly tested (current test coverage is above 90%)
on Python version >= 3.5+ and 2.7 (as of 2020, support for Python2 is
discontinued). Although automatic continuous integration (CI) systems are not
in place, we do our best to regularly tests under new Python versions, when
available. Remember that tests are time consuming (some minutes currently).
Here some examples depending on your needs:
pytest -xvvv -W ignore ./tests/
pytest -xvvv -W ignore --dburl postgresql://<user>:<password>@localhost/<dbname> ./tests/
pytest -xvvv -W ignore --dburl postgresql://<user>:<password>@localhost/<dbname> --cov=./stream2segment --cov-report=html ./tests/
Where the options denote:
-x: stop at first error-vvv: increase verbosity,-W ignore: do not print Python warnings issued during tests. You can omit the-Woption to turn warnings on and inspect them, but consider that a lot of redundant messages will be printed: in case of test failure, it is hard to spot the relevant error message. Alternatively, try-W once- warn once per process - and-W module-warn once per calling module.--cov: track code coverage, to know how much code has been executed during tests, and--cov-report: type of report (if html, you will have to opend 'index.html' in the project directory 'htmlcov')--dburl: Additional database to use. The default database is an in-memory sqlite database (e.g., no file will be created), thus this option is basically for testing the program also on postgres. In the example, the postgres is installed locally (localhost) but it does not need to. Remember that a database with name<dbname>must be created first in postgres, and that the data in any given postgres database will be overwritten if not empty
-
(update January 2021) On MacOS (version 11.1, with Python 3.8 and 3.9):
-
if the installation fails with a lot of printout and you spot a "Failed building wheel for psycopg2", try to execute:
export LIBRARY_PATH=$LIBRARY_PATH:/usr/local/opt/openssl/lib/ && pip ./installme-dev(you might need to change the path of openssl below). Credits here and here)
-
If the error message is "Failed building wheel for tables", then
brew install c-bloscand re-runinstallme-devinstallation command (with theexportcommand above, if needed)
-
-
If you see (we experienced this while running tests, thus we can guess you should see it whenever accessing the program for the first time):
This system supports the C.UTF-8 locale which is recommended. You might be able to resolve your issue by exporting the following environment variables: export LC_ALL=C.UTF-8 export LANG=C.UTF-8Then edit your
~/.profile(or~/.bash_profileon Mac) and put the two lines starting with 'export', and executesource ~/.profile(source ~/.bash_profileon Mac) and re-execute the program. -
On Ubuntu 12.10, there might be problems with libxml (
version libxml2_2.9.0' not found). Move the file or create a link in the proper folder. The problem has been solved looking at http://phersung.blogspot.de/2013/06/how-to-compile-libxml2-for-lxml-python.html
All following issues should be solved by installing all dependencies as described in the section Prerequisites. If you did not install them, here the solutions to common problems you might have and that we collected from several Ubuntu installations:
-
For numpy installation problems (such as
Cannot compile 'Python.h') , the fix has been to update gcc and install python3-dev (python2.7-dev if you are using Python2.7, discouraged):sudo apt-get update sudo apt-get upgrade gcc sudo apt-get install python3-devFor details see here
-
For scipy problems,
build-essential gfortran libatlas-base-devare required for scipy. For details see here -
For lxml problems,
libxml2-dev libxslt-devare required. For details see here -
For matplotlib problems (matplotlib is not used by the program but from imported libraries),
libpng-dev libfreetype6-devare required. For details see here and here
-
Although PEP8 recommends 79 character length, the program used initially a 100 characters max line width, which is being reverted to 79 (you might see mixed lengths in the modules). It seems that among new features planned for Python 4 there is an increment to 89.5 characters. If true, we might stick to that in the future
-
In the absence of Continuous Integration in place, from times to times, it is necessary to update the dependencies, to make
pip installmore likely to work (at least for some time). The procedure is:pip install -e . pip freeze > ./requirements.tmp pip install -e ".[dev]" pip freeze > ./requirements.dev.tmp(you could also do it with jupyter). Remember to comment the line of stream2segment from each requirements (as it should be installed as argument of pip:
pip install <options> ., and not inside the requirements file). Run tests (see above) with warnings on: fix what might go wrong, and eventually you can replace the oldrequirements.txtandrequirements.dev.txtwith the.tmpfile created. -
Updating wiki:
The wiki is the documentation of this project accessible on the 'wki' tab on the GitHub page. It is composed of markdown (.md) files (one file per page), some written manually and some generated from the notebooks (.ipynb) of this project via the
jupyter nbconvertcommand on the terminal: currently, the files 'The-Segment-object' and 'Using-Stream2segment-in-your-Python-code'. Consider implementing a script (see the draft 'resources/templates/create_wiki.py') if the amount of pages increases considerably.To update the wiki (update existing notebooks or add new one):
Requirements:
jupyterinstalled.- A clone of the repo 'stream2segment.wiki' on the same parent directory of the stream2segment repo.
Then:
- Edit the notebooks in stream2segment/resources/templates:
- Some new notebook? First choose a meaningful file name, as it will be the title of the wiki page (hyphens will be replaced with spaces in the titles). Then check that the notebook is tested, i.e. run in "tests/misc/test_notebook"
- Create .md (markdown) versions of the notebook for the wiki. From the
stream2segment repository as
cwd(Fis the filename without the 'ipnyb' extension):(repeat for every notebook file, e.g.F='Using-Stream2segment-in-your-Python-code';jupyter nbconvert --to markdown ./stream2segment/resources/templates/$F.ipynb --output-dir ../stream2segment.wiki
The-Segment-object) cd ../stream2segment.wiki:- Some new notebook? add it in the TOC of 'Home.md' if needed, and
git add - Eventually,
git commit,pushas usual. Check online. Done
- Some new notebook? add it in the TOC of 'Home.md' if needed, and