AIMD data analysis methods and part of the raw data for the paper "Supramolecular Proton Conductors Self-Assembled by Organic Cages".
View contents on website: Molecular-Dynamics-Molecular-Cages
The analysis methods are described in the md_ana.ipynb file. You can also view the contents on web page: Detailed methods.
The analysis results are available in the cage1 and cage2 folders. The files in the above folders are:
-
sys-pos-1.xyzthe AIMD raw trajectory file dumped by CP2K code run on Archer2. -
cage*_500K.xyzthe trajectory file calibrated by fix the centre of mass of the system. -
water_com_cage*.xlsxthe centre of masses of water molecules -
MSD_D_H2O_cage*.xlsxthe mean square displacements and diffusion coefficients of water molecules. -
hbonds_mic.jsonthe hydrogen bond analysis results, including the number of H-bonds in each frame, the donor, acceptor, the donor-acceptor separation, the D-H···A angle. For cage-2, only 3000 frames are included due to file size limit. -
Figures:
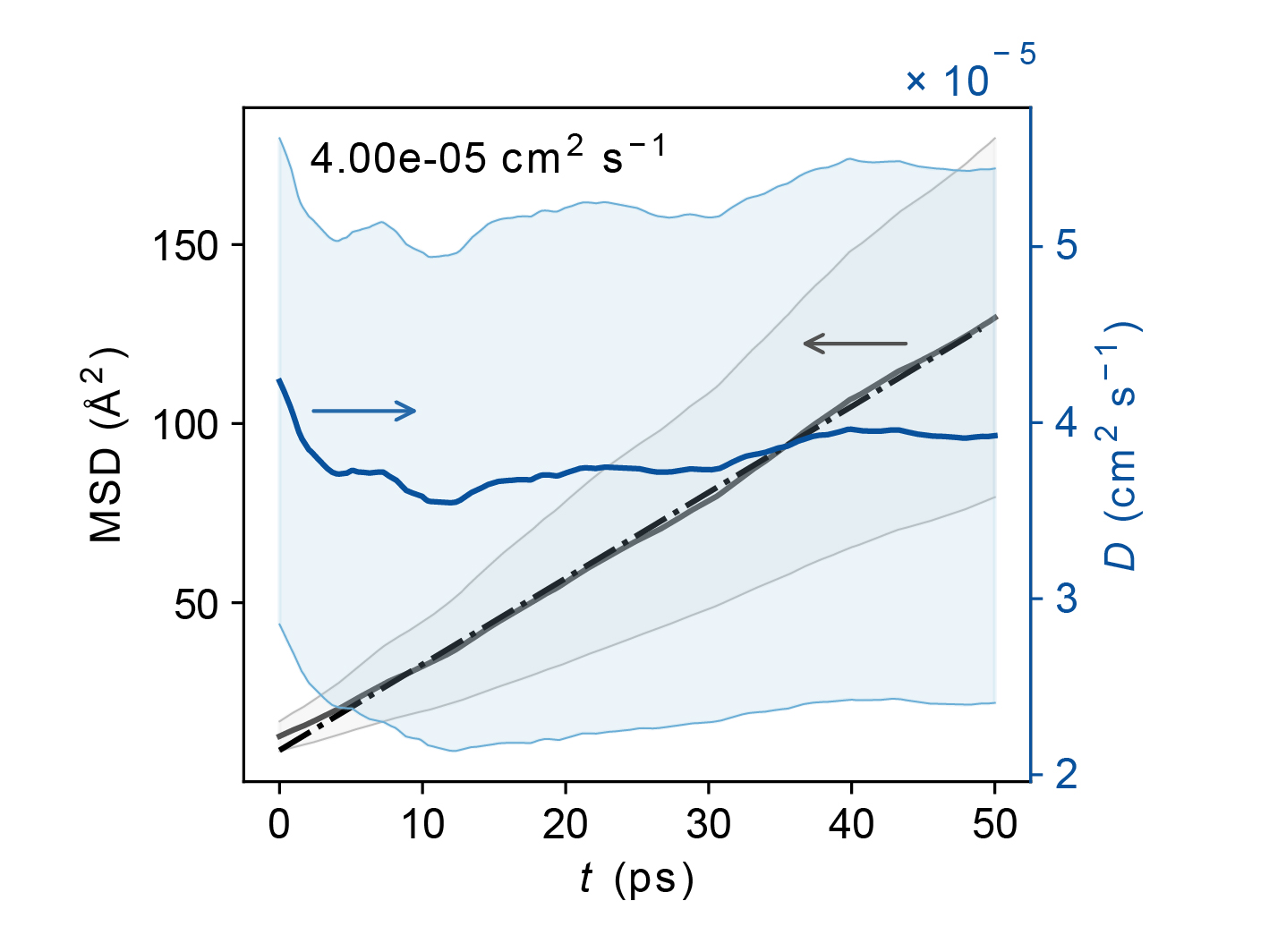
cage*_500K_ave.jpgAverage MSD and diffusion coefficient with error bounds.
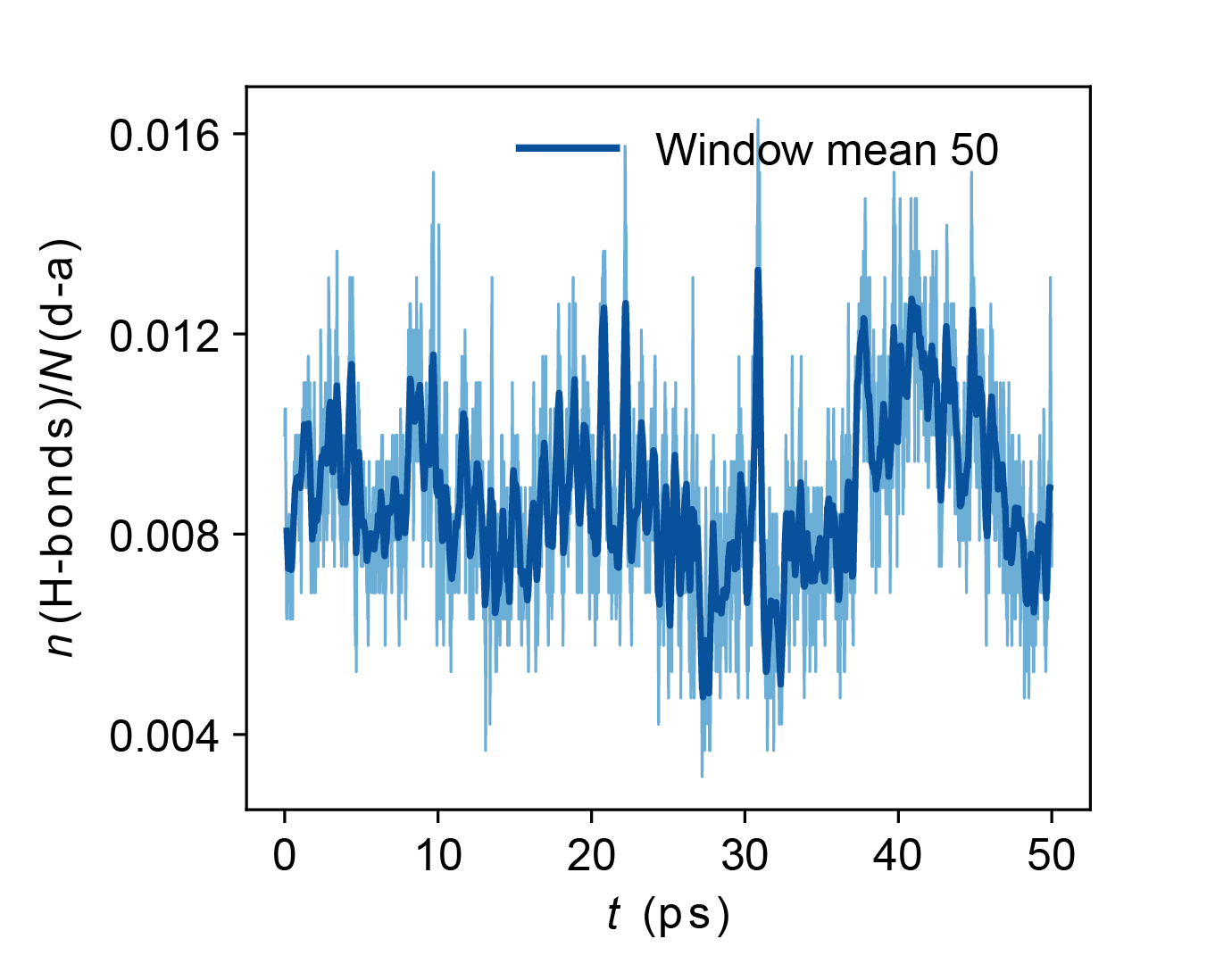
cage*_nhbonds_500K.jpgNumber of Hydrogen bonds per unique hydrogen bonding donor-acceptor pairs with respect to time.
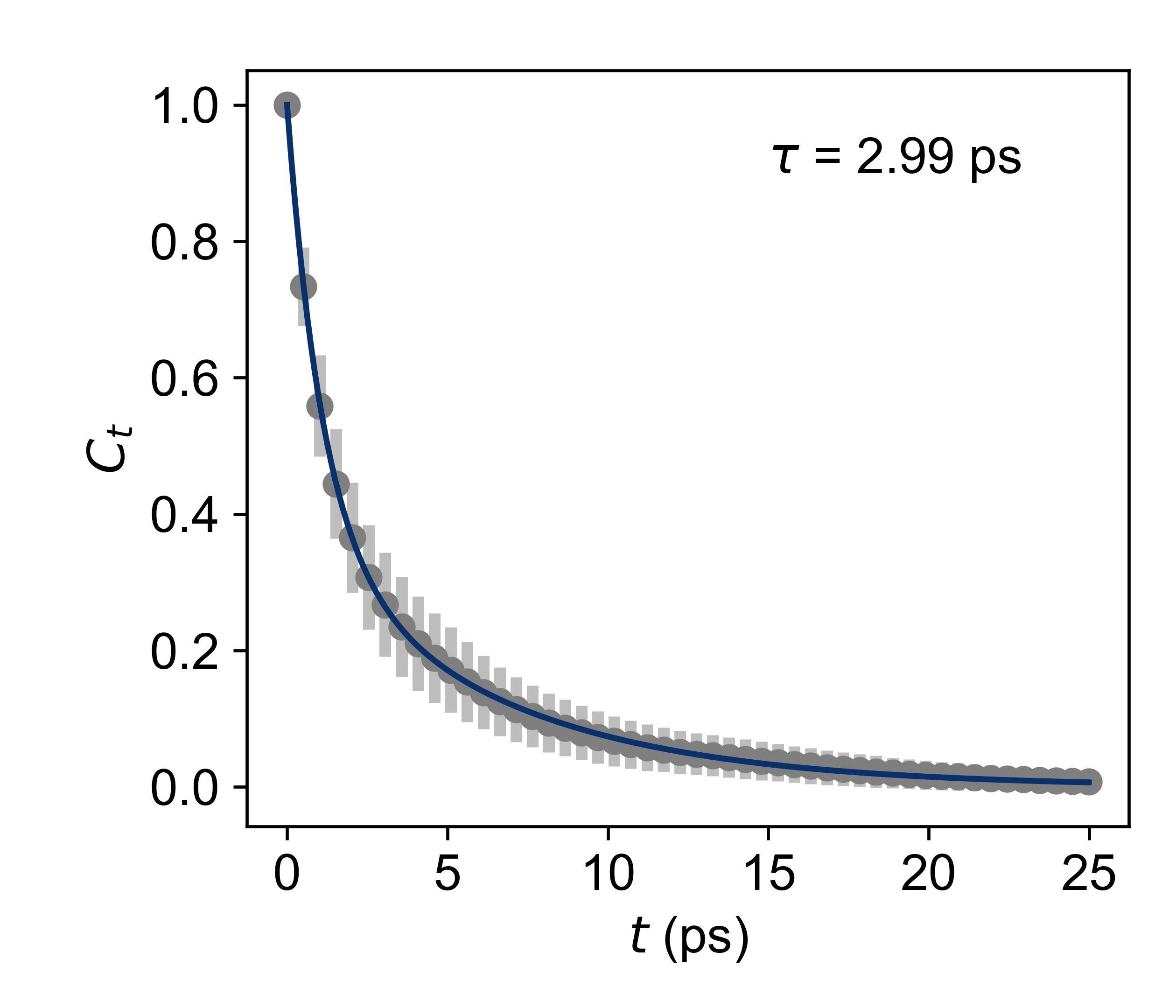
cage*_hbond_lifetime_500K.jpgHydrogen bond lifetime.
The raw .xyz trajectory files were dumped for every 5 fs (every 10 AIMD steps), and are large (several hundreds of MB). Part of the raw trajectory data (650 frames obtained by evenly extracting one frame per every 15 frames from the raw data, the time interval between two consecutive frames is 75 fs) have been uploaded to the cage1 and cage2 folders. For our analysis, all frames (over 11,000 frames, > 55,000 fs or 55 ps) in the large raw trajectory files were used. The raw trajectory files can be provided in reasonable request to reproduce our results.
Our paper was published on JACS Au: Zhenyu Yang, Ningjin Zhang, Lei Lei, Chunyang Yu, Junjie Ding, Pan Li, Jiaolong Chen, Ming Li, Sanliang Ling, Xiaodong Zhuang, and Shaodong Zhang, JACS Au 2022 2 (4), 819-826. Please ensure you cite our paper if you used our methods in your research.
Please contact Lei Lei or Dr Sanliang Ling for further computational details of the simulation results. Any other correspondence regarding the paper should be sent to Dr Shaodong Zhang.