This repository contains resources and information related to the SARS-CoV-2 ARTIC Panel for Illumina. The primers in this set are based on the published, improved sequences (version N1) by Itokawa et. al 2020.
See this this github repository for more information and the original sequences.
The repository is split into two parts:
├── primers
└── example_workflow
- primers
- Contains a file with primer sequences, binding positions and amplicion ranges with respect to the SARS-Cov2 MN908947.3 genome assembly.
- example_workflow
- Features a showcase script and some resources on how one could perform analysis of the obtained data.
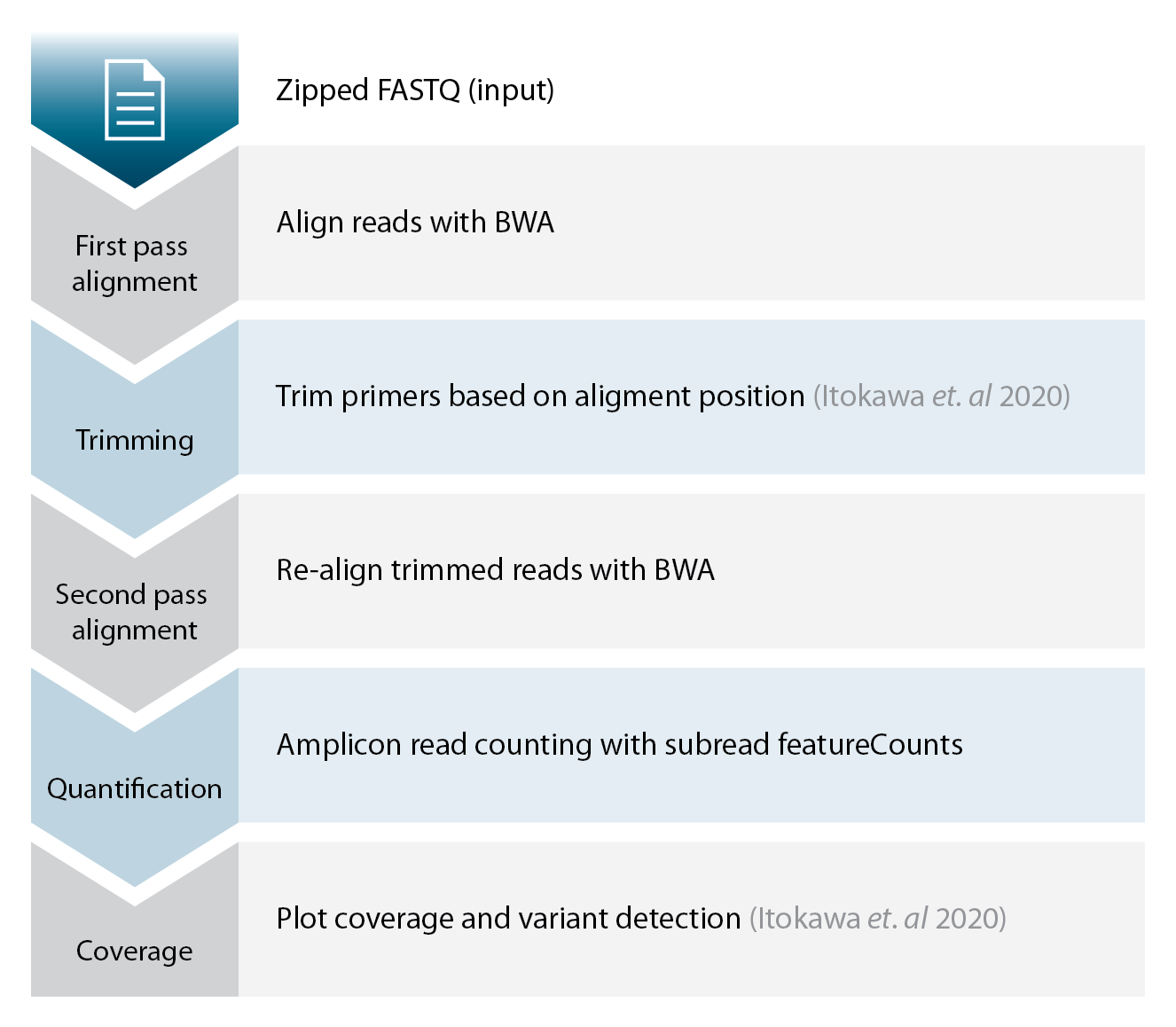
As mentioned above, the example_workflow folder contains an example script as well as serveral other resources that will make it easier to get you started with your data analysis. This script implements the following steps and is based among others on tools and a workflow published by Itokawa et. al 2020.
The basic steps of the analysis are outlined below. There is no need for adapter trimming if you use Illumina sequencers. This is because the shortest amplicon should be 380 bp, while Illumina sequencers only allow paired-end sequencing up to 300 bp. The workflow does include a position based primer trimming step though. If you are using our unique dual indicies for barcoding also check out our demultiplexing tool.
The example_workflow directory contains the following files or folders:
example_workflow
├── environment.yml
├── example_workflow.sh
├── fc_annotation.saf
└── MN908947.3
- environment.yml
- A yaml file to install the required dependencies into a conda environment.
- This can easily be done by running the command
conda env create -f environment.yml - See here for an excellent tutorial on what conda is.
- example_workflow.sh
- The above shown example workflow is implemented in bash
- The script contains several variables at the beginning that might need to be edited.
- If you want to run this script make sure you downloaded the required scripts from this github repository.
- fc_annotation.saf
- A simple annotation format file that can be used to quantify the amplicons with featureCounts
- This file has been generated from the orginal resource published by Itokawa et. al 2020. See here for the original file.
- MN908947.3
- A folder containing the MN908947.3 genome assembly indexed for BWA 0.7
-
Itokawa K, Sekizuka T, Hashino M, Tanaka R, Kuroda M (2020) Disentangling primer interactions improves SARS-CoV-2 genome sequencing by multiplex tiling PCR. PLoS ONE 15(9): e0239403
-
Tools by Itokawa et. al used by the example workflow - https://github.com/ItokawaK/Alt_nCov2019_primers
-
The original ARTIC primer panel - https://github.com/artic-network/artic-ncov2019
-
SARS-Cov2 genome assembly and annotation (MN908947.3)