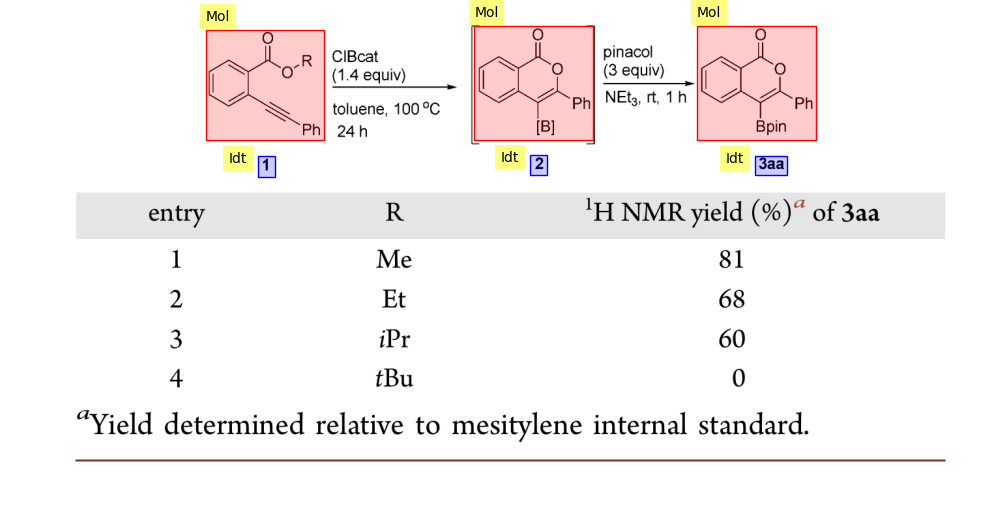
This is the repository for MolDetect and MolCoref, two sequence generation models for reaction diagram parsing.
Run the following command to install the package and its dependencies:
git clone git@github.com:Ozymandias314/MolDetect.git
cd RxnScribe
python setup.py install
Download the checkpoint and use MolDetect to extract molecules from reaction diagrams:
import torch
from rxnscribe import MolDetect
from huggingface_hub import hf_hub_download
ckpt_path = hf_hub_download("Ozymandias314/MolDetectCkpt", "best_hf.ckpt")
model = MolDetect(ckpt_path, device=torch.device('cpu'))
image_file = "assets/jacs.5b12989-Table-c3.png"
predictions = model.predict_image_file(image_file)The predictions will be in the following format:
[
{ #first bbox
'category': '[Sup]',
'bbox': (0.0050025012506253125, 0.38273870663142223, 0.9934967483741871, 0.9450094869920168),
'category_id': 4,
'score': -0.07593922317028046
},
#More bounding boxes
]We provide a function to visualize the predicted bboxes:
visualize_images = model.draw_bboxes(predictions, image_file = image_file)Each predicted diagram will be visualized in a seperate image, where red boxes are molecules, green boxes are text, blue boxes are identifiers. gold boxes are supplementary information.
To detect bounding boxes and also the coreference, download the coreference checkpoint for MolDetect on huggingface and also set the coref field to be true:
import torch
from rxnscribe import MolDetect
from huggingface_hub import hf_hub_download
ckpt_path = hf_hub_download("Ozymandias314/MolDetectCkpt", "coref_best_hf.ckpt")
model = MolDetect(ckpt_path, device=torch.device('cpu'), coref = True)
image_file = "assets/jacs.5b12989-Table-c3.png"
predictions = model.predict_image_file(image_file, coref = True)The predictions will be in the following format:
{
'bboxes': [
{ #first bbox
'category': '[Sup]',
'bbox': (0.0050025012506253125, 0.38273870663142223, 0.9934967483741871, 0.9450094869920168),
'category_id': 4,
'score': -0.07593922317028046
},
#More bounding boxes
],
'coref': [
([0, 1], "4a"), #coreferences contain the indices of the molecule and identifier bounding boxes, as well as the string representation of the identifier
([3, 4], "4b"),
#More coref pairs
]
}Similarly, we provide a function to visualize the predicted bboxes:
visualize_images = model.draw_bboxes(predictions, image_file = image_file, coref = True)Each predicted diagram will be visualized in a seperate image, where each molecule-coreference pair is depicted with the same color.
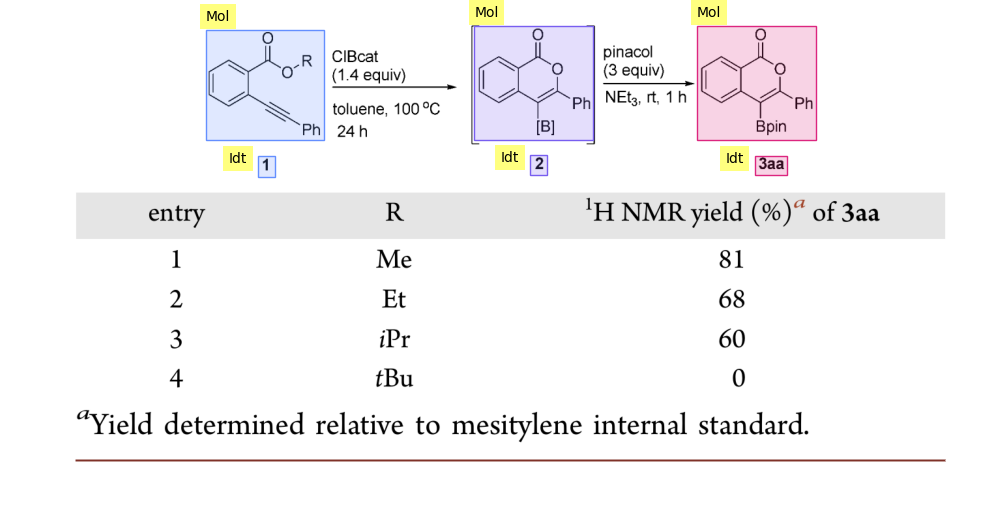
For development or reproducing the experiments, follow the instructions below.
Install the required packages
pip install -r requirements.txt
Download the reaction diagrams from this link,
and save them to data/detect/images/.
The ground truth files can be found at data/detect/splits/annotations/ and data/coref/splits/annotations/.
We use a 70-10-20 split in our experiments. The full train/dev/test split is available in the above directories.
This notebook shows how to visualize the diagram and the ground truth.
Run this script to train and evaluate MolDetect on our train test split.
bash scripts/train_detect.shRun this script to train and evaluate MolCoref on our train test split.
bash scripts/train_coref.sh