Multi-omics integration for both single-cell and spatially resolved data based on dual-path graph attention auto-encoder
Single-cell multi-omics integration enables joint analysis at the single-cell level of resolution to provide more accurate understanding of complex biological systems, while spatial multi-omics integration is benefit to the exploration of cell spatial heterogeneity to facilitate more comprehensive downstream analyses. Existing methods are mainly designed for single-cell multi-omics data with little consideration on spatial information, and still have the room for performance improvement. A reliable multi-omics integration method designed for both single-cell and spatially resolved data is necessary and significant. We propose a multi-omics integration method based on dual-path graph attention auto-encoder (SSGATE). It can construct the neighborhood graphs based on single-cell expression profiles or spatial coordinates, enabling it to process single-cell data and utilize spatial information from spatially resolved data. It can also perform self-supervised learning for integration through the graph attention auto-encoders from two paths. SSGATE is applied to integration of transcriptomics and proteomics, including single-cell and spatially resolved data of various tissues from different sequencing technologies. SSGATE shows better performance and stronger robustness than competitive methods and facilitates downstream analysis.
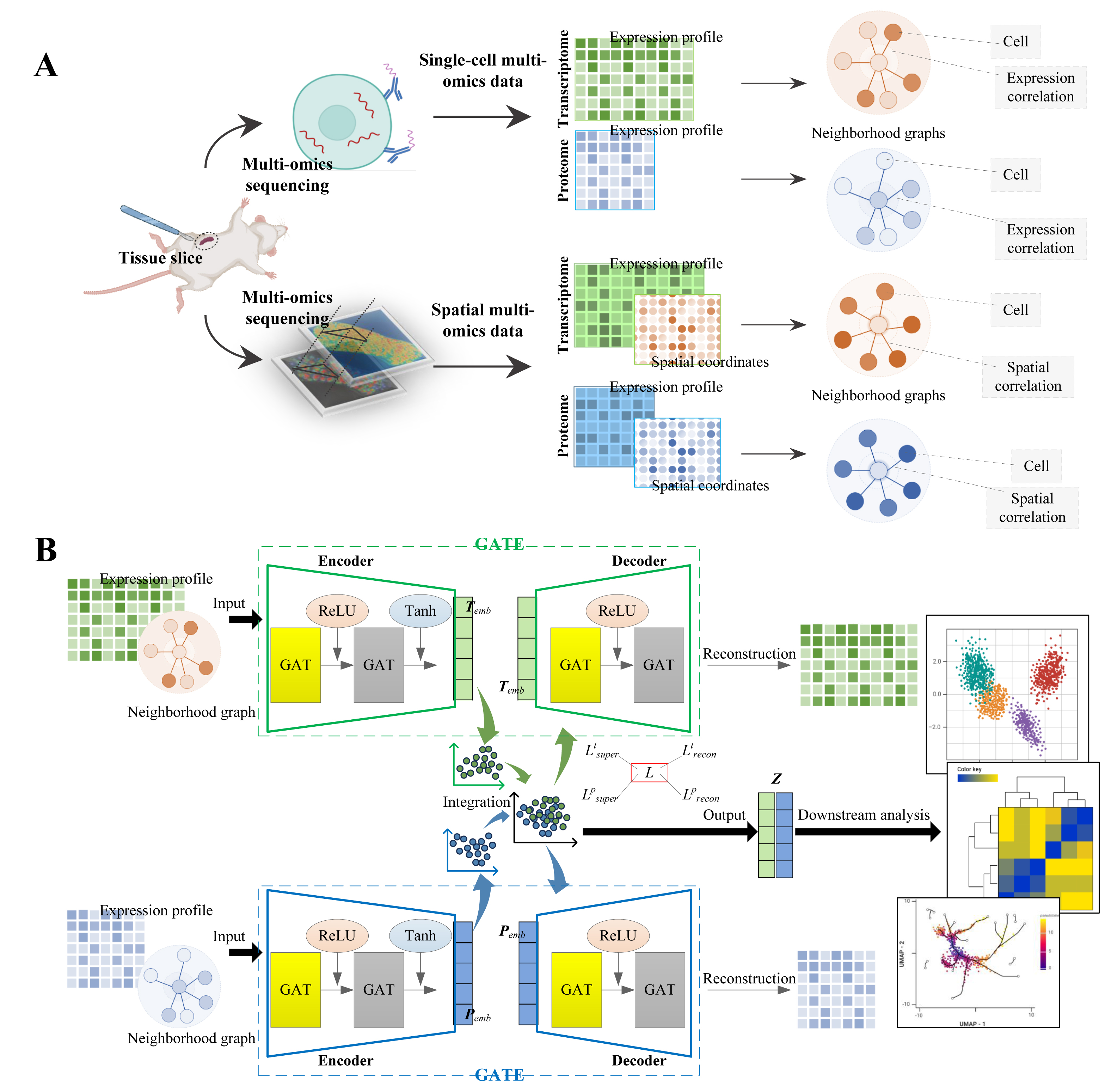
Figure 1. Overview of SSGATE. (A) Neighborhood graph construction for single-cell multi-omics and spatial multi-omics data. (B) Dual-path graph attention auto-encoder for multi-omics integration.
Download this GitHub repository, and extract the contents into a folder.
Note: SSGATE requires data in anndata format as inputs.
The expression profiles are required to be stored in anndata.X, otherwise the data cannot be read
correctly. The spatial coordinates from spatial multi-omics data are required to be stored in
anndata.obsm['spatial'], otherwise it will be processed as single-cell multi-omics data due to
the spatial information cannot be read.
We provide the datasets used in this study, which are stored in the BGI cloud disk. Link Extract password is :nrEB
### Python enviroment constructed by Conda
conda create -n SSGATE python=3.9
conda activate SSGATE
git clone https://github.com/Linliu-Bioinf/SSGATE.git
cd SSGATE
python setup.py install
pip install -r requirements.txtWe recommend using STOmics Cloud Platform to access and use it.
Users can use the public image of the STOmics Cloud Platform: SSGATE. The dependency files related to
running SSGATE have been configured.
Before running SSGATE for multi-omics joint feature analysis, you must use the Cal_Nbrs_Net function to construct a neighbor
network for a single omics data (transcriptome or proteome).
adata = Cal_Nbrs_Net(adata,
feat='X_pca',
rad_cutoff=None,
k_cutoff=None,
model='Radius',
verbose=True):
The parameters of Cal_Nbrs_Net are:
adata: An AnnData object, where obsm contains spatial coordinates or coordinate information.feat: The feature matrix used to calculate the neighbors. Can be either reduced coordinates ('X_pca') or spatial coordinates ('spatial').rad_cutoff: When using the radius neighbor model, defines the neighborhood radius. If using the Radius model, this value must be provided.k_cutoff: Defines the number of neighbors when using the KNN model. This value is required if the KNN model is used.model: Specifies the type of model to use. Possible values are 'Radius' and 'KNN'.verbose: If True, the function will print progress information as it calculates.
return value:
adata: The updated AnnData object, containing the calculated neighboring network information, is stored in adata.uns['nbrs_net'].
This function is used to prune the adjacency network calculated by the Cal_Nbrs_Net function, ensuring that each edge
only connects cells within the same cell population. After pruning, the updated adjacency network will be stored in
the uns attribute of the AnnData object.
adata = prune_net(adata)
return value:
adata: The updated AnnData object, containing the pruned neighbor network information, is stored in uns['nbrs_net'].
This function is used to prune the adjacency network calculated by the Cal_Nbrs_Net function, ensuring that each edge
only connects cells within the same cell population. After pruning, the updated adjacency network will be stored in
the uns attribute of the AnnData object.
adata_st, adata_sp = ssmi.train(adata_st,
adata_sp,
hidden_dims1 = 128,
hidden_dims2 = 128,
out_dims = 30,
cluster_update_epoch = 20,
epochs_init = 50,
n_epochs=300,
lr=0.001,
save_reconstrction=False,
sigma = 0.1,
device = "cuda:0",
feat1 = "PCA")
The parameters of train are:
adata_st: An AnnData object containing transcriptomics data.adata_sp: AnnData object containing proteomics data.hidden_dims1: Hidden layer dimensions for the first dataset in the model.hidden_dims2: Hidden layer dimensions for the second dataset in the model.out_dims: The dimensions of the model output embeddings.cluster_update_epoch: The interval between updating cluster labels.epochs_init: Number of epochs in the initial training phase.n_epochs: The total number of training rounds.lr: Learning rate.key_added: The key under which the embed result is stored.gradient_clipping: Gradient clipping threshold.weight_decay: Weight decay coefficient.verbose: Controls the output of training process information.random_seed: Random seed to ensure reproducibility.save_loss: Whether to save training loss.save_reconstrction: Whether to save the reconstruction results.sigma: The weight of the triplet loss in the loss function.margin: Margin of triplet loss.device: The training device (GPU or CPU).feat1: Features used by the first dataset.feat2: Features used by the second dataset.
We provide a basic tutorial on using SSGATE (Tutorial_of_SSGATE.ipynb).
If you use SSGATE in your work, please cite it.
Single-cell multi-omics and spatial multi-omics data integration via dual-path graph attention auto-encoder
Tongxuan Lv, Yong Zhang, Junlin Liu, Qiang Kang, Lin Liu
bioRxiv 2024 June 04. doi: https://doi.org/10.1101/2024.06.03.597266.
Any questions or suggestions on SSGATE are welcomed! Please report it on issues, or contact Lin Liu (liulin4@genomics.cn).