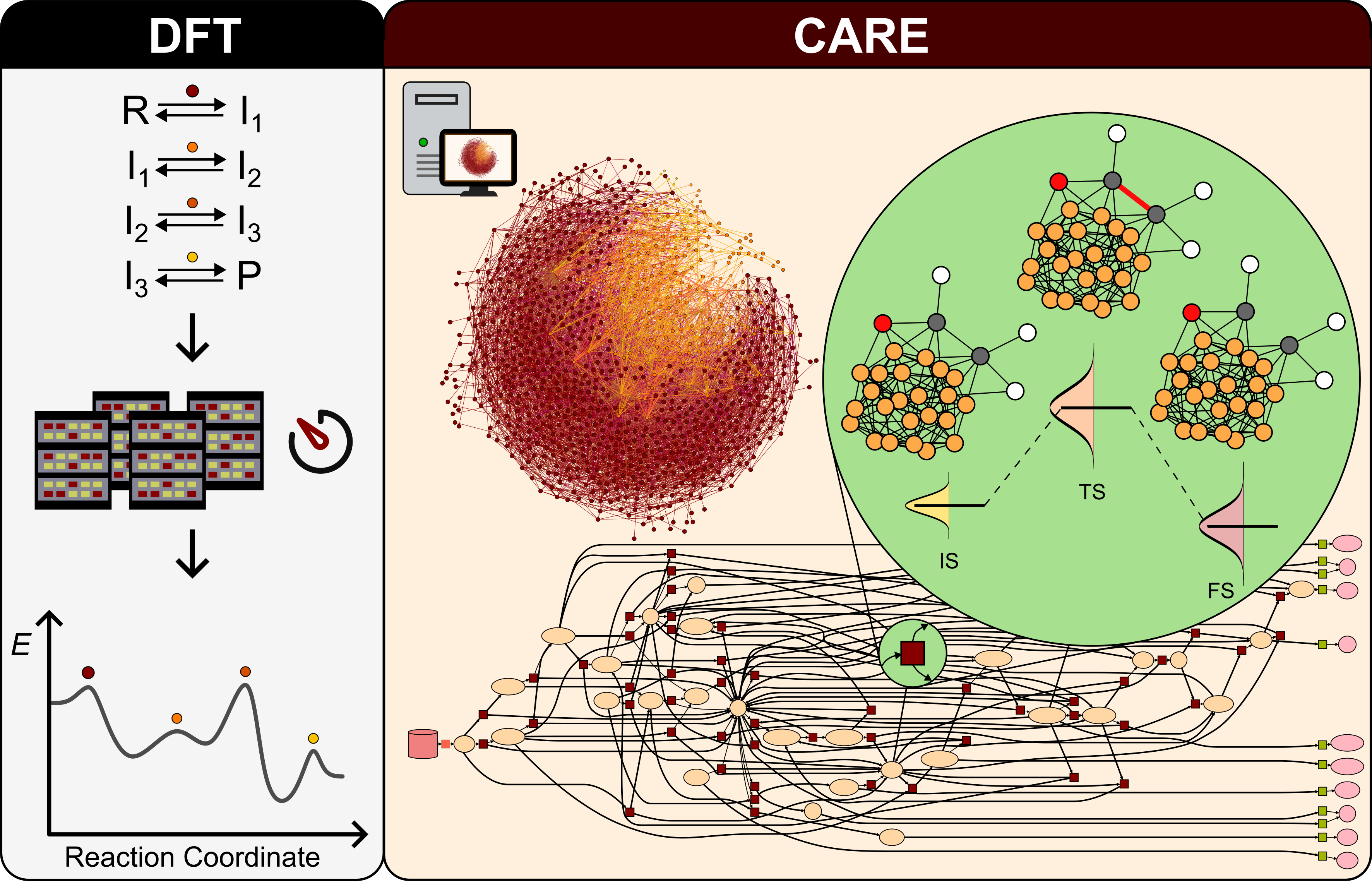
CARE (Catalysis Automated Reaction Evaluator) is a tool for generating and manipulating chemical reaction networks (CRNs) on catalytic surfaces. CARE is powered with GAME-Net-UQ, a graph neural network with uncertainty quantification targeting the DFT energy of relaxed species and transition states.
Installing CARE requires Conda and Git locally installed. The following instructions are optimized to install CARE on Linux systems, while for macOS we noticed a lower performance in the CRN generation mainly due to Python multiprocessing (see Contexts and start methods in the documentation)
- Clone the repo:
git clone git@github.com:LopezGroup-ICIQ/care.git- Create a conda environment with Python 3.11 and activate it:
conda create -n care_env python=3.11
conda activate care_env- Enter the repo and install the package with pip:
cd care
python3 -m pip install .NOTE: MacOS users might need to launch a new shell at this point in order for the entry points to work correctly.
- Install
pytorchandpytorch_geometricthrough conda. Ensure that the libraries are obtained from the pytorch and pyg channels, as shown here:
conda install pytorch cpuonly pytorch-scatter pytorch-sparse pyg -c pytorch -c pygNOTE: MacOS users might need to install pytorch geometric using pip.
- (optional) Install Julia and the ODE packages required to perform microkinetic simulations. As alternative, simulations can run with the implemented Scipy solver.
curl -fsSL https://install.julialang.org | sh
python3 -m pip install juliacall # Python-Julia bridge
julia -e 'import Pkg; Pkg.add("DifferentialEquations"); Pkg.add("DiffEqGPU"); Pkg.add("CUDA");'NOTE: For some systems Julia may present some error while using sh. If that is the case, please install Julia by running instead:
curl -fsSL https://install.julialang.org | sh -s -- -y- (optional) Install the different evaluators available (MACE, fairchem), through the following command:
NOTE: There currently is a dependency clash during installation for the two evaluators related to the e3nn library (see: this issue for MACE). Installation might result in an incompatibility warning, but
both libraries should work correctly if the installation order shown below is followed.
python3 -m pip install fairchem-core
python3 -m pip install mace-torchgen_crn_blueprint -h # documentation
gen_crn_blueprint [-ncc NCC] [-noc NOC] [-electro ELECTRO] [-o OUTPUT] [-ncpu NUM_CPU]This script does not require any input file. The output is stored as pickle file. To access the blueprint, do:
from pickle import load
with open('path_to_blueprint_file', 'rb') as f:
intermediates, reactions = load(f)eval_crn -h # documentation
eval_crn [-i INPUT] [-bp BP] [-o OUTPUT] [-ncpu NUM_CPU]This script requires an input toml file defining the surface of interest, property evaluators, and their settings. The output is a ReactionNetwork object stored as pickle file. You can find an example input file here.
run_kinetic [-i INPUT] [-crn CRN] [-o OUTPUT]This script runs microkinetic simulation starting from the evaluated reaction network and an input toml file defining the reaction conditions, solver, inlet conditions. The results are stored as a pickle object file.
You can run the entire pipeline (blueprint generation -> properties evaluation -> kinetic simulation) running the care_run script:
care_run -h # documentation
care_run -i input.toml -o output_nameThis will generate a directory output_name containing a crn.pkl with the generated reaction network.
Examples of input .toml files can be found in src/care/scripts and src/care/examples.
We currently provide three tutorials, available in the notebooks directory:
The DFT database in ASE format used to retrieve available CRN intermediates will be uploaded soon in Zenodo.
The code is released under the MIT license.
- A Foundational Model for Reaction Networks on Metal Surfaces Authors: S. Morandi, O. Loveday, T. Renningholtz, S. Pablo-García, R. A. Vargas Hernáńdez, R. R. Seemakurthi, P. Sanz Berman, R. García-Muelas, A. Aspuru-Guzik, and N. López DOI: 10.26434/chemrxiv-2024-bfv3d