3D-Unet: patched based Pytorch implementation for medical images segmentation
Important News -- Repository Maintenance
This repository will no longer be developed and improved. A new version for medical images segmentation is available at https://github.com/davidiommi/SALMON
3D-Unet pipeline is a computational toolbox for segmentation using neural networks.
Same script applied for prostate segmentation can be found here: https://github.com/davidiommi/3D_Prostate_segmentation_pytorch
The training and the inference are patch based: the script randomly extract corresponding patches of the images and labels and feed them to the network during training. The inference script extract, segment the sigle patches and automatically recontruct them in the original size.
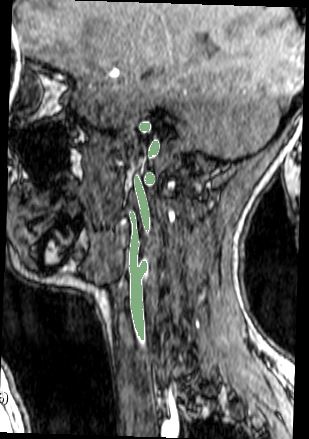
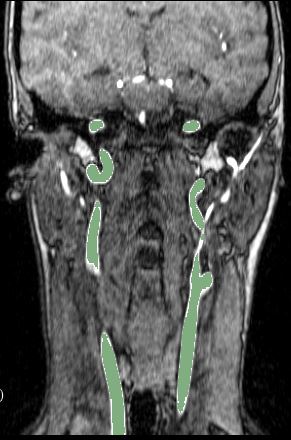
Example images
Sample MR images from the sagittal and coronal views for carotid artery segmentation (the segmentation result is highlighted in green)
Requirements
pillow scikit-learn simpleITK keras scikit-image pandas pydicom nibabel tqdm torch>=0.4.1 torchvision>=0.2.1 dominate>=2.3.1 visdom>=0.1.8.
Python scripts and their function
-
organize_folder_structure.py: Organize the data in the folder structure for the network
-
NiftiDataset.py : They augment the data, extract the patches and feed them to the network (reads .nii files). NiftiDataset.py skeleton taken from https://github.com/jackyko1991/unet3d-pytorch
-
check_loader_patches: Shows example of patches fed to the network during the training
-
UNet.py: the architecture of the U-net.
-
utils.py : list of metrics and loss functions for the training
-
main.py: Runs the training and the prediction on the training and validation dataset.
-
predict.py: It launches the inference on training and validation data in the main.py
-
predict_single_image.py: It launches the inference on a single input image chosen by the user.
Usage
Use first organize_folder_structure.py to create organize the data in the following folder structure Modify the init.py to set the parameters and start the training/testing on the data:
Folder Structure:
.
├── Data_folder
| ├── train_set
| | ├── patient_1 # Training
| | | ├── image # Contains domain image
| | | └── label # Contains domain label
| | └── patient_2
| | | ├── image
| | | └── label
| ├── test_set
| | ├── patient_3 # Testing
| | | ├── image
| | | └── label
| | └── patient_4
| | | ├── image
| | | └── label
| ├── validation_set
| | ├── patient_5 # Validation
| | | ├── image
| | | └── label
| | └── patient_6
| | | ├── image
| | | └── label
Features
- 3D data processing ready
- Augmented patching technique, requires less image input for training
- one channel output (multichannel to be developed)
- Generic image reader with SimpleITK support (Currently only support .nii/.nii.gz format for convenience, easy to expand to DICOM, tiff and jpg format)
- Medical image pre-post processing with SimpleITK filters
- Easy network replacement structure
- Dice score similarity measurement as golden standard in medical image segmentation benchmarking