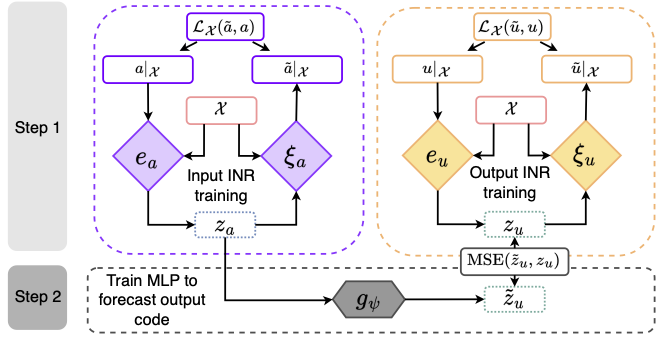
Official PyTorch implementation of CORAL | Accepted at Neurips 2023 | Arxiv
To cite our work:
@article{serrano2023operator,
title={Operator Learning with Neural Fields: Tackling PDEs on General Geometries},
author={Louis Serrano and Lise Le Boudec and Armand Kassaï Koupaï and Thomas X Wang and Yuan Yin and Jean-Noël Vittaut and Patrick Gallinari},
journal={37th Conference on Neural Information Processing Systems (NeurIPS 2023)},
year={2023},
url={https://arxiv.org/abs/2306.07266}
}
conda create -n coral python=3.9.0
pip install -e .
pip install torch_geometric
pip install pyg_lib torch_scatter torch_sparse torch_cluster torch_spline_conv -f https://data.pyg.org/whl/torch-2.0.0+cu117.html
add to your ~/.bashrc
export WANDB_API_TOKEN=your_key
export WANDB_DIR=your_dir
export WANDB_CACHE_DIR=your_cache_dir
export WANDB_CONFIG_DIR="${WANDB_DIR}config/wandb"
export MINICONDA_PATH=your_anaconda_path
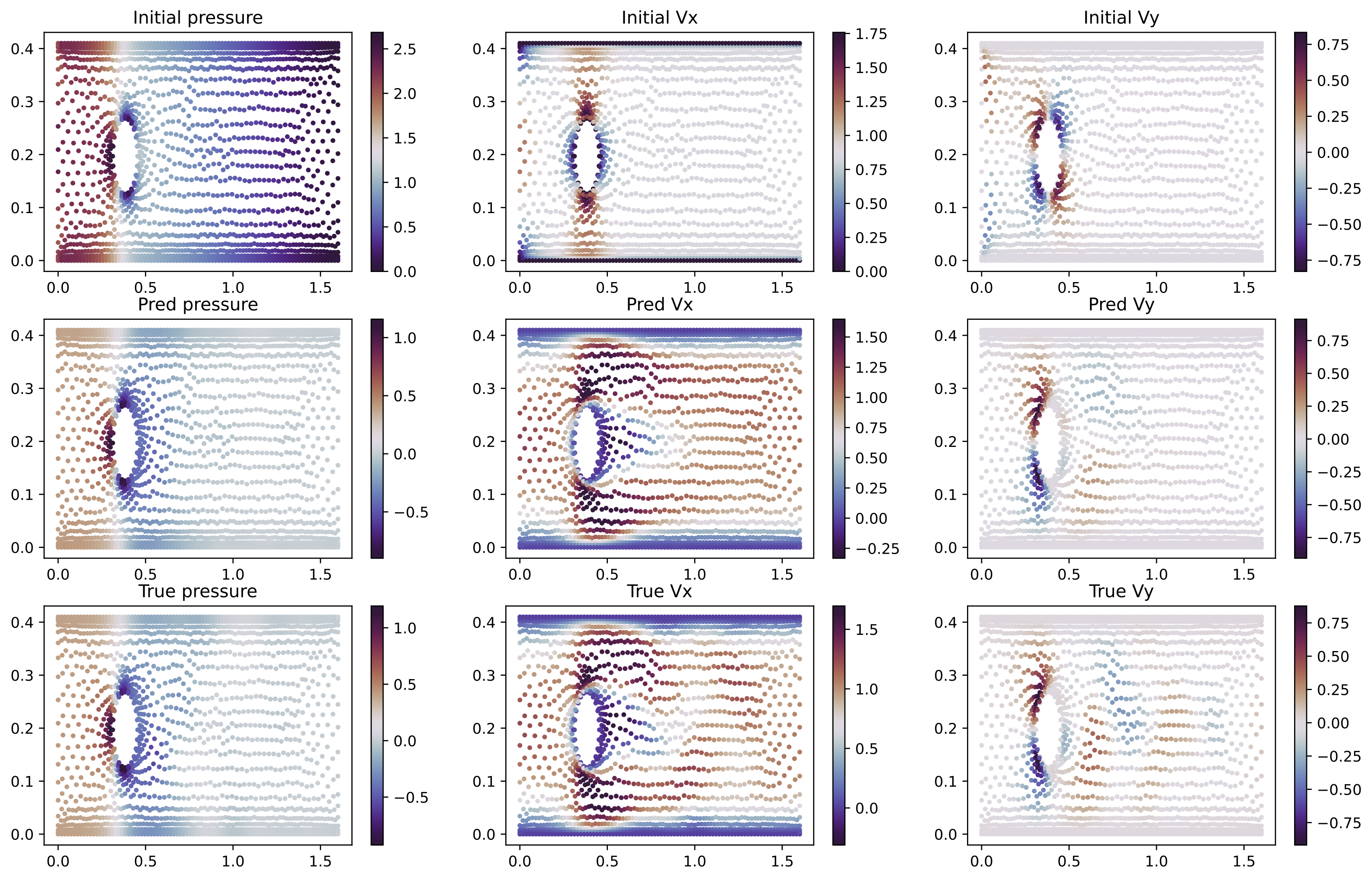
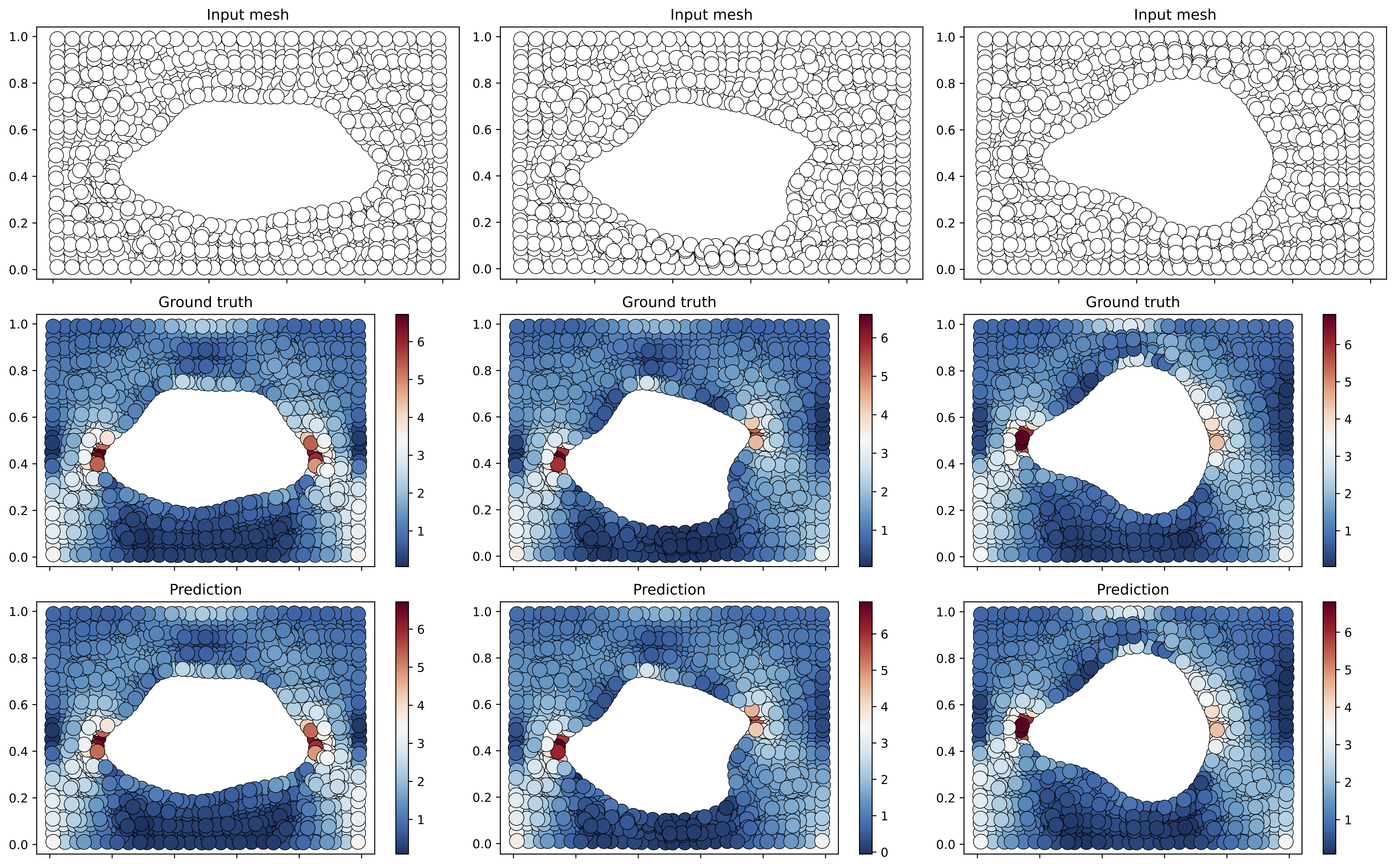
- IVP: Cylinder and Airfoil datasets can be downloaded from : https://github.com/deepmind/deepmind-research/tree/master/meshgraphnets
- Dynamics Modeling: NS and SW data can be generated using https://github.com/mkirchmeyer/DINo (SW data accessible here after issues raised by wanghonghui1998 : https://drive.google.com/drive/folders/1RlH4E7-dnKOEA6Zur2EdK-HuEaW_PC8w?usp=sharing)
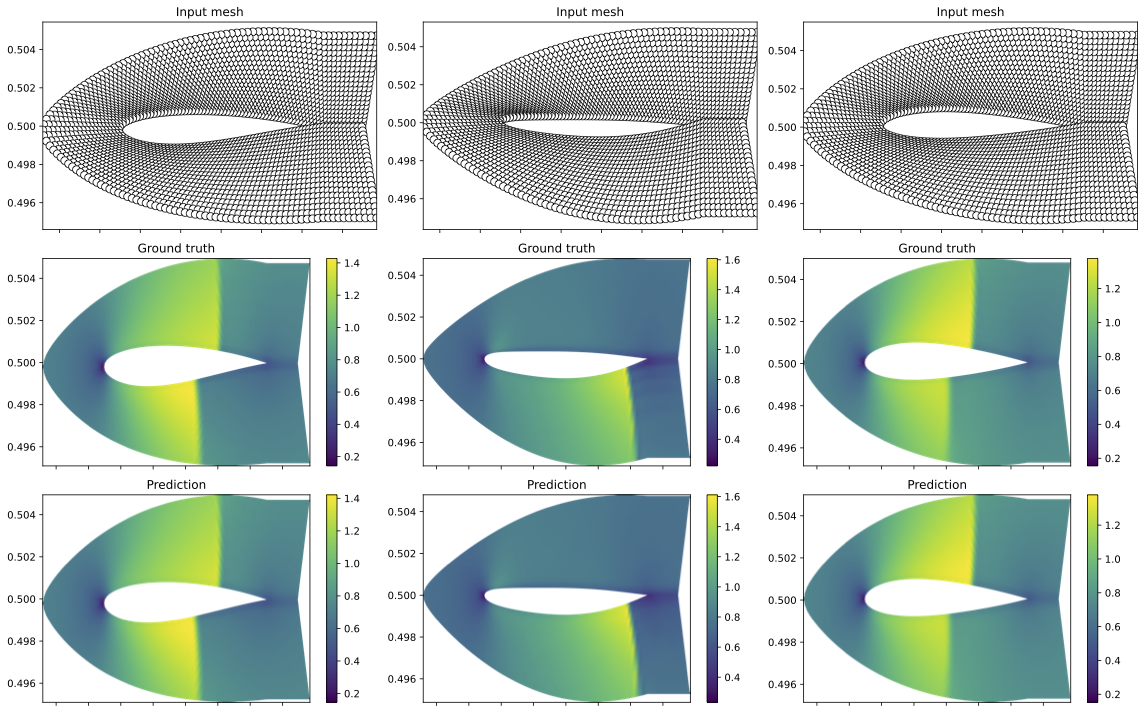
- Geo-FNO: NACA-Euler, Elasticity and pipe data can be downloaded from https://drive.google.com/drive/folders/1YBuaoTdOSr_qzaow-G-iwvbUI7fiUzu8 (Note that NACA-Euler stands for Airfoil in this google drive)
The code runs only on GPU. We provide sbatch configuration files to run the training scripts. They are located in bash_static and bash_dynamics.
We expect the user to have wandb installed in its environment to ease the 2-step training.
For all tasks, the first step is to launch an inr.py training. The weights of the inr model are automatically saved under its run_name.
For the second step, i.e. for training the dynamics or inference model, we need to use the previous run_name as input to the config file to load the inr model.
We provide examples of the python scripts that need to be run.
- airfoil-flow:
python3 static/train/ivp_inr.py "data.dataset_name=airfoil-flow" 'optim.epochs=5000' 'inr_in.w0=5' 'inr_out.w0=5' 'inr_in.latent_dim=128' 'inr_in.hidden_dim=256' 'inr_out.hidden_dim=256' 'inr_out.latent_dim=128' 'model.width=256' 'optim.batch_size=32' 'optim.meta_lr_code=5e-6'
python3 static/train/ivp_regression.py "data.dataset_name=airfoil-flow" "inr.run_name=dandy-lion-4438" "optim.epochs=101"
- cylinder-flow:
python3 static/train/ivp_inr.py "data.dataset_name=cylinder-flow" 'optim.epochs=5000' 'inr_in.w0=20' 'inr_out.w0=15' 'inr_in.latent_dim=128' 'inr_out.latent_dim=128' 'optim.epochs=2000' 'model.width=64'
python3 static/train/ivp_regression.py "data.dataset_name=cylinder-flow" "inr.run_name=dandy-puddle-3918" "optim.epochs=2001"
python3 static/train/design_inr.py "data.dataset_name=airfoil" 'optim.epochs=5000' 'inr_in.w0=5' 'inr_out.w0=15' 'optim.lr_inr=1e-4' 'optim.meta_lr_code=1e-4'
python3 static/train/design_regression.py "data.dataset_name=airfoil" "inr.run_name=glowing-music-4181" 'optim.epochs=10000'
- elasticity
python3 static/train/design_inr.py "data.dataset_name=elasticity" 'optim.batch_size=64' 'optim.epochs=5000' 'inr_in.w0=10' 'inr_out.w0=15' 'optim.lr_inr=1e-4' 'optim.meta_lr_code=1e-4'
python3 static/train/design_regression.py "data.dataset_name=elasticity" "inr.run_name=clone-nerf-herder-4289" 'optim.epochs=10000'
- pipe
python3 static/train/design_inr.py "data.dataset_name=pipe" 'optim.batch_size=16' 'optim.epochs=5000' 'inr_in.w0=5' 'inr_out.w0=10' 'inr_in.hidden_dim=128' 'inr_in.depth=5' 'inr_out.hidden_dim=128' 'inr_out.depth=5' 'optim.lr_inr=5e-5' 'optim.meta_lr_code=5e-5'
python3 static/train/design_regression.py "data.dataset_name=pipe" "inr.run_name=super-plasma-4149" 'optim.epochs=10000' 'model.width=128' 'model.depth=3' 'inr.inner_steps=3'
python3 inr/inr.py "data.sub_from=$sub_from" "data.same_grid=$same_grid" "data.dataset_name=$dataset_name" "inr.model_type=$model_type" "data.seq_inter_len=$seq_inter_len" "data.seq_extra_len=$seq_extra_len" "data.sub_tr=$sub_tr" "data.sub_te=$sub_te" "optim.lr_inr=$lr_inr" "inr.depth=$depth" "inr.latent_dim=$latent_dim" "inr.hidden_dim=$hidden_dim" "optim.batch_size=$batch_size" "optim.epochs=$epochs" "wandb.saved_checkpoint=$saved_checkpoint" #"wandb.name=$name" "wandb.id=$id" "wandb.dir=$dir" "wandb.checkpoint_path=$checkpoint_path"
python3 dynamics_modeling/train.py "data.sub_from=$sub_from" "data.same_grid=$same_grid" "data.dataset_name=$dataset_name" "dynamics.width=$width" "dynamics.depth=$depth" "data.sub_tr=$sub_tr" "data.sub_te=$sub_te" "optim.epochs=$epochs" "data.seq_inter_len=$seq_inter_len" "data.seq_extra_len=$seq_extra_len" "optim.batch_size=$batch_size" "optim.lr=$lr" "dynamics.teacher_forcing_update=$teacher_forcing_update" "inr.run_name=$run_name"
- shallow-water:
python3 inr/inr.py "data.sub_from=$sub_from" "data.same_grid=$same_grid" "data.dataset_name=$dataset_name" "data.data_to_encode=$data_to_encode" "data.seq_inter_len=$seq_inter_len" "data.seq_extra_len=$seq_extra_len" "data.sub_tr=$sub_tr" "data.sub_te=$sub_te" "optim.lr_inr=$lr_inr" "inr.depth=$depth" "inr.latent_dim=$latent_dim" "inr.hidden_dim=$hidden_dim" "optim.batch_size=$batch_size" "inr.w0=$w0" "optim.epochs=$epochs" "wandb.saved_checkpoint=$saved_checkpoint" #"wandb.name=$name" "wandb.id=$id" "wandb.dir=$dir" "wandb.checkpoint_path=$checkpoint_path"
python3 dynamics_modeling/train.py "data.sub_from=$sub_from" "data.same_grid=$same_grid" "data.dataset_name=$dataset_name" "dynamics.width=$width" "dynamics.depth=$depth" "data.sub_tr=$sub_tr" "data.sub_te=$sub_te" "optim.epochs=$epochs" "data.seq_inter_len=$seq_inter_len" "data.seq_extra_len=$seq_extra_len" "optim.batch_size=$batch_size" "optim.lr=$lr" "dynamics.teacher_forcing_update=$teacher_forcing_update" "inr.run_dict=$run_dict"
with dataset_name='navier-stokes-dino' or 'shallow-water-dino'.