This repository provides a pipeline to compare the result of EM algorithm and LDSC algorithm in the context of GWAS. The input data is the UK Biobank data for height and the output is the comparison of the heritability estimation by EM and LDSC. The pipeline includes the following steps:
The document of EM Algorithm can be found in EM and the document of LDSC can be found in LDSC.
- Calculate LDScore: Calculate the LD score
- Generate_Simulation_Data: Generate the simulation data
- Run EM: Run EM algorithm
- Run LDSC in py: Run LDSC algorithm in python
- Run LDSC in R: Run LDSC algorithm in R
- Visualize: Visualize the result
- Run All in Server: Run all the above in server
- shell: Shell scripts to run the above sequentially (or separately) in server
The Usage of each script is described in the header of each script. You can set parameters in the GLOBAL SETTINGS part of run.slurm to run the pipeline. Or follow the following steps to run the pipeline step by step.
Here's a script to run all the above. Remember to change the settings in GlobalSettings.sh accordingly.
cd to the directory of the code scripts, then run the following script:
calLd_id=$(sbatch --parsable shell/CalculateLDScore.sh)
gendata_id=$(sbatch --parsable --dependency=afterok:$calLd_id shell/GenerateData.sh)
em_id=$(sbatch --parsable --dependency=afterok:$gendata_id shell/EM.sh)
ldsc_id=$(sbatch --parsable --dependency=afterok:$gendata_id shell/LDSC.sh)
sbatch --dependency=afterok:$em_id:$ldsc_id shell/Visual.sh
- Upload the UKBTNNI plink data (.bed, .bim, .fam) to server and change the setting in GlobalSettings.sh accordingly.
The structure of the files should be look like this:
> tree ${data_path} -L 1
.
├── height_ukb_50k_TNNI1.bed
├── height_ukb_50k_TNNI1.bim
├── height_ukb_50k_TNNI1.fam
├── ...
- cd to the directory of the code scripts, for example:
cd /home/wjiang49/UKBheight/
The slurm log file will be saved in the above directory like this:
log
├── slurm_out_614489.log
├── slurm_out_614490.log
├── slurm_out_614491.log
├── slurm_out_615299_1.log
├── slurm_out_615299_2.log
├── slurm_out_615299_3.log
- Calculate LD score and record the job id for the following steps:
calLd_id=$(sbatch --parsable shell/CalculateLDScore.sh)
The .ldscore file will be saved in the same directory as the plink data.
This will calculate the LD score and save the result in the same directory as the plink data.
- Generate the simulation data:
gendata_id=$(sbatch --parsable --dependency=afterok:$calLd_id shell/GenerateData.sh)
The results will be saved in the ${output_path} directory. If such directory does not exist, it will be created. The generated data shall look like this:
.
├── genotypes.csv
├── h0.1
...
├── h0.9
└── log
The summary data and phenotype data will be saved in the h0.* directory. The log of all the process will be saved in the log directory.
- Run EM and LDSC. The job will begin after the generation of the simulation data. The log will be saved in the
${output_path}/logdirectory and results will be saved in the${output_path}directory.
i. Run EM:
em_id=$(sbatch --parsable --dependency=afterok:$gendata_id shell/EM.sh)
ii. Run LDSC:
ldsc_id=$(sbatch --parsable --dependency=afterok:$gendata_id shell/LDSC.sh)
After that, the ${output_path} directory should look like this:
.
├── em_results.csv
├── genotypes.csv
├── h0.1
...
├── h0.9
├── ldsc_results.csv
└── log
- Visualize the result:
sbatch --dependency=afterok:$em_id:$ldsc_id shell/Visual.sh
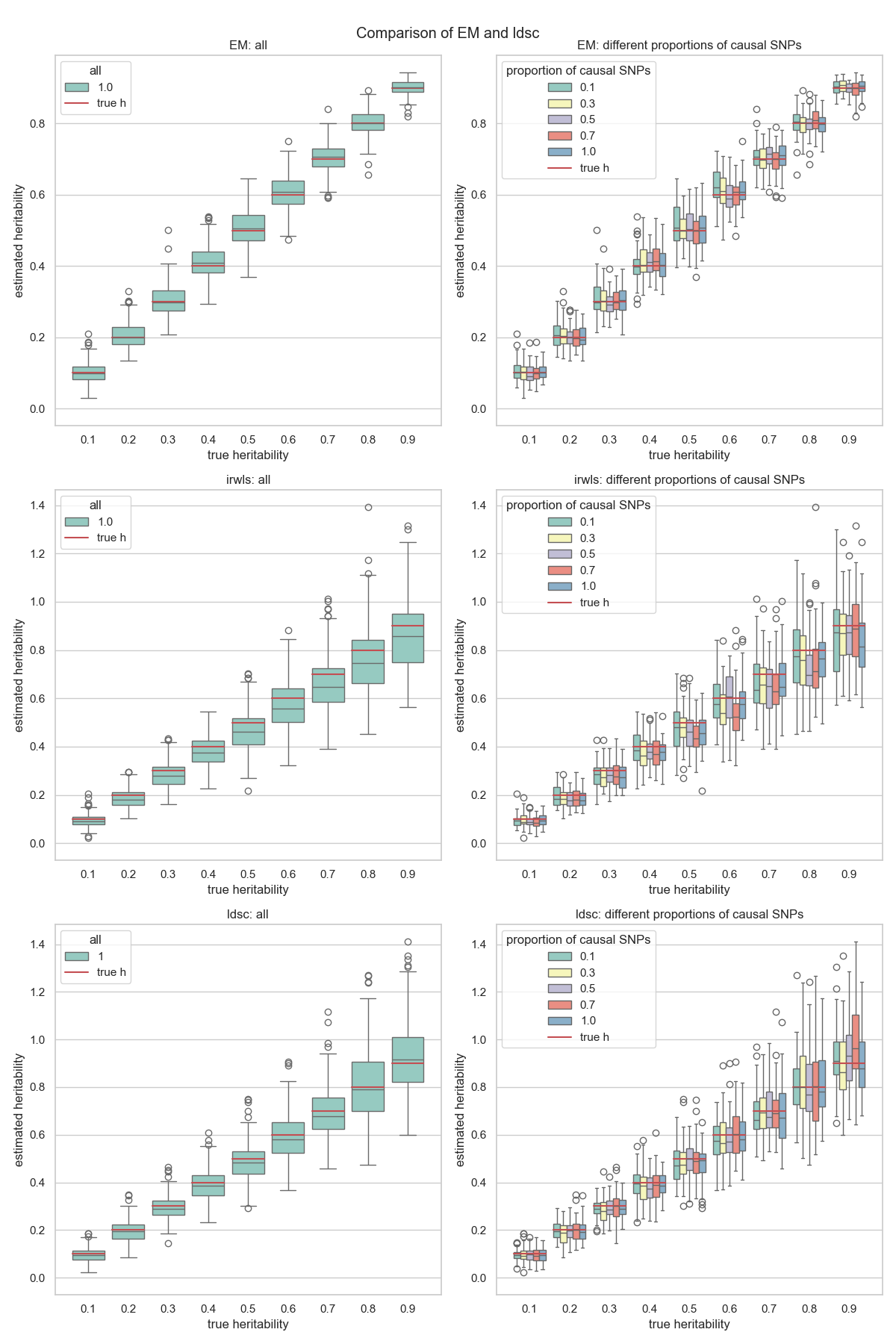
Also, the figures will be saved in the ${output_path} directory. You can get a figure like this:
- The running status can be checked by:
>squeue -p special_bios
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
623813 special_b UKBHeigh wjiang49 PD 0:00 1 (Dependency)
623812_[1-3] special_b UKBHeigh wjiang49 PD 0:00 1 (Dependency)
623814 special_b UKBHeigh wjiang49 PD 0:00 1 (Dependency)
623809_1 special_b UKBHeigh wjiang49 R 1:59 1 hpc-gpu007
623809_2 special_b UKBHeigh wjiang49 R 1:59 1 hpc-gpu007
623809_3 special_b UKBHeigh wjiang49 R 1:59 1 hpc-gpu007
END