Quick workaround for this bug/feature request: snakemake/snakemake#660 Use at your own risk.
$ python setup.py installThe tool outputs a shell command which removes a list of file. The command is
python -m snakemake_workaround rm-command --rule <my rule> --node0 <src node> --node1 <des node> --plot <True/False>
where:
<my rule>is a rule which builds a dag in your Snakemake pipeline, like the rule you want to execute- each of
<src node>and<des node>can either be a full-path, refering to one of the input/output of your pipeline, either a rule- if one
<src node>is specified, the list of files in output is made of all the files that are "downstream" the node - if both
<src node>and<des node>are specified, the list of files in output is made of all the files that are "between" the two nodes.
- if one
- the boolean flag for plotting shows what is going on, by highlighting the list of files given as output
Example command:
rm-command --rule short --node0 compute_region_centers --node1 _train_models --plot True
Red nodes are rules, green nodes are files, black nodes aggregates list of inputs for the same rule, and make visualization a bit easier.
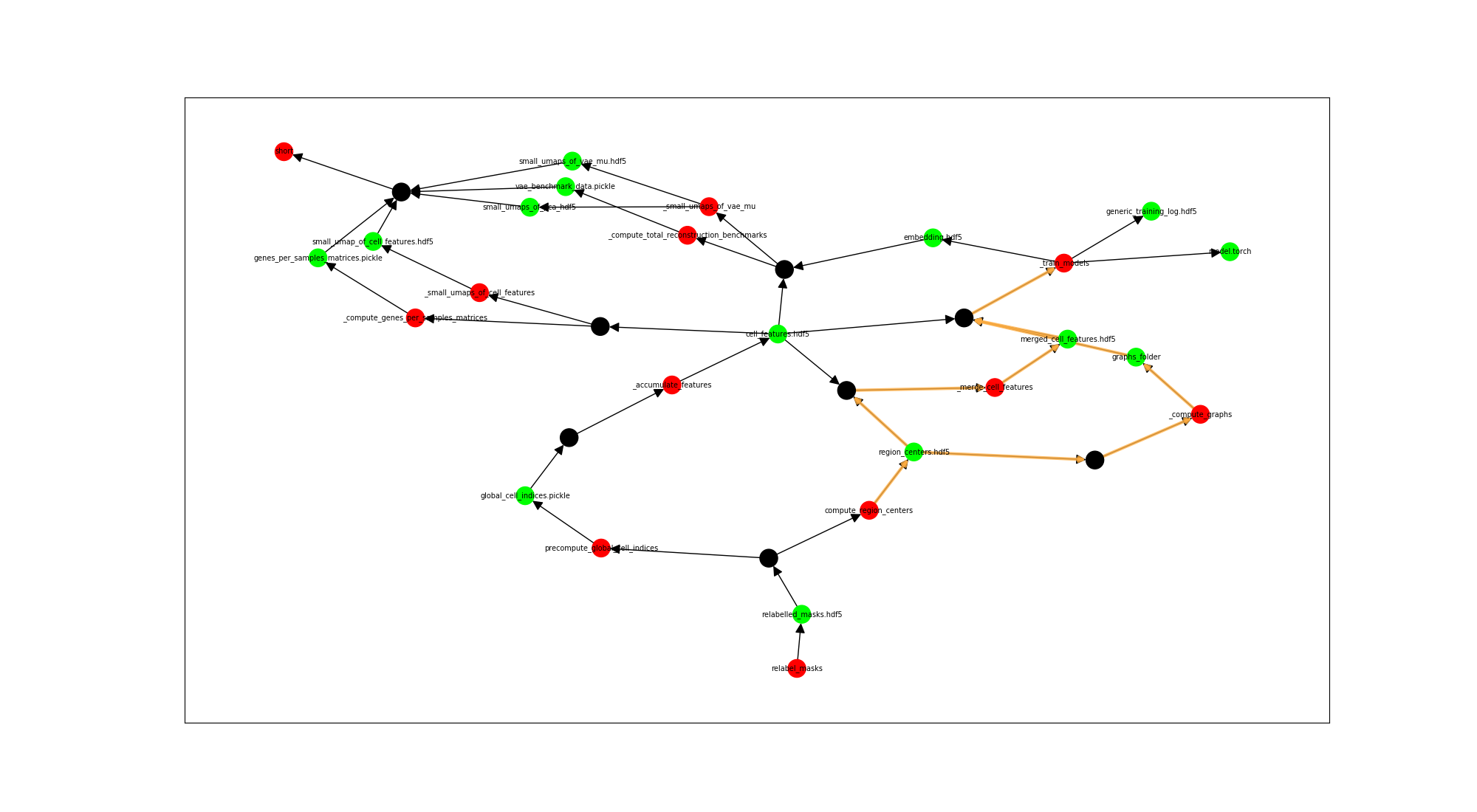
Example output:
for f in /data/l989o/spatial_uzh/data/spatial_uzh_processed/region_centers.hdf5 /data/l989o/spatial_uzh/data/spatial_uzh_processed/phyper_data/graphs/0f867a8979f43b5b384434989df1a8b6929fce8cd78d95f6ff1d184d844d7983/graphs_folder/ /data/l989o/spatial_uzh/data/spatial_uzh_processed/phyper_data/accumulated_features/4522b0aa174e0f8c8a5890fc31da5feb032d2c3163211aba373e995273629f6d/merged_cell_features.hdf5; do rm $f; done
This package was created with Cookiecutter and the oldani/cookiecutter-simple-pypackage project template.