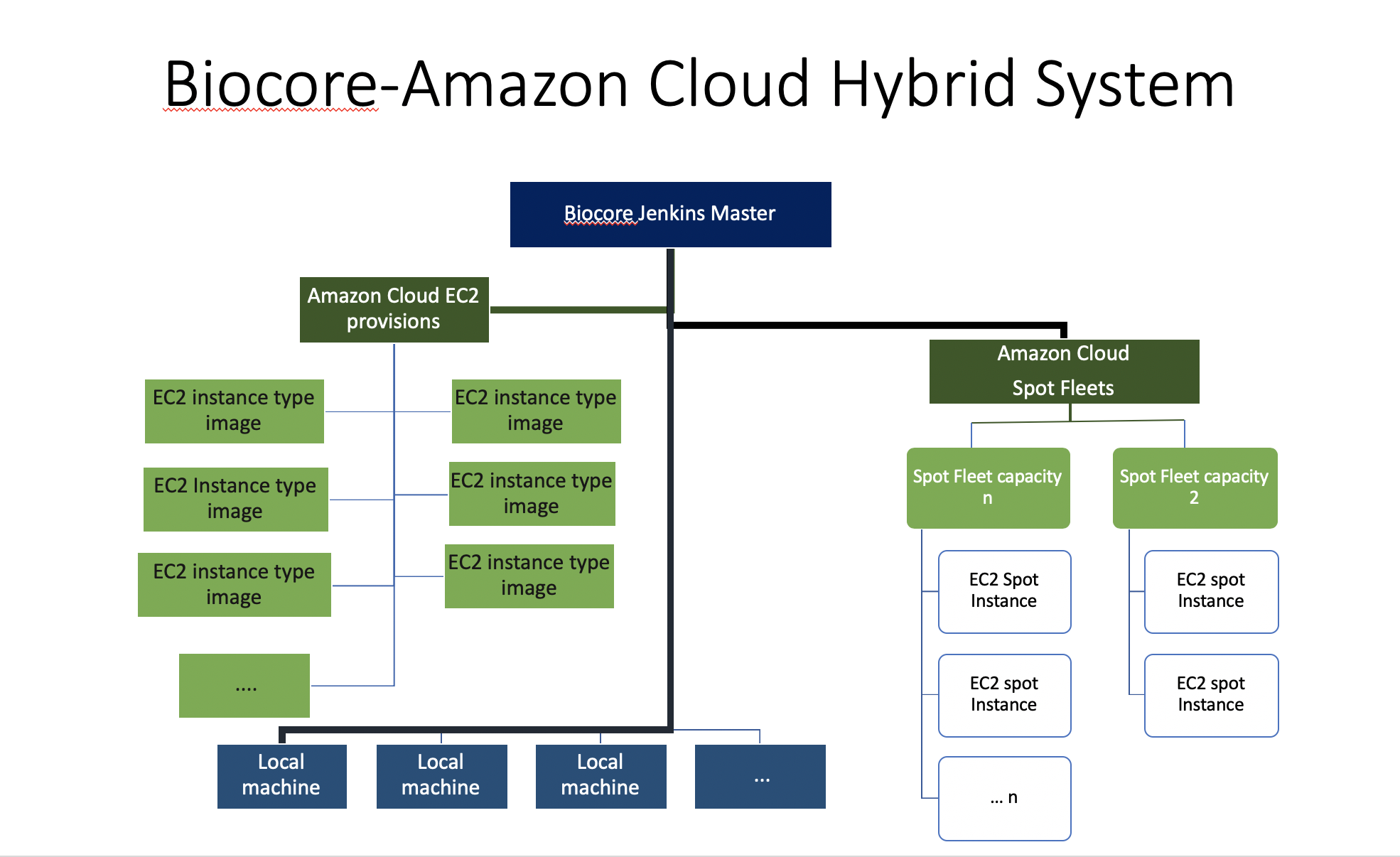
Biocore uses Jenkins to launch pipeline analysis projects either on local servers or on the Amazon Cloud servers, or on both. Jenkins integrates very well across languages, platforms, and operating systems - Additionally Jenkins is widely documented and open-source software.
Our Jenkins setting consists on A single master server with multiple worker nodes. Worker nodes are a hybrid of Amazon EC2 instances and MDIBL servers
- Biocore-AWS Hybrid System Overview
- Biocore Pipeline Run Options And Steps
- Repository Information Organization
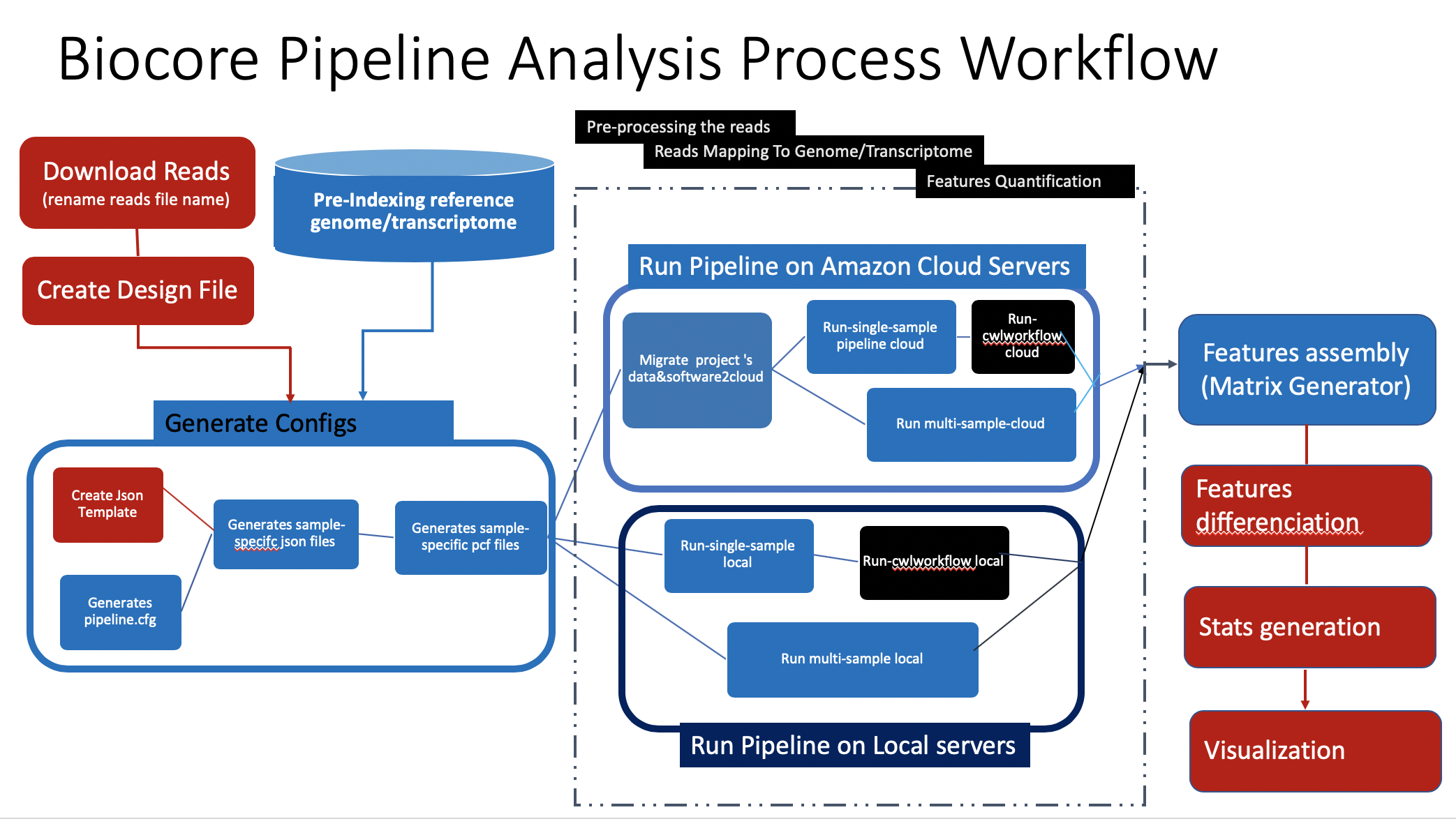
Depending of the workload, you have these options to run your pipeline :
The root directory is organized as followed:
A set of biocore global settings and program-specific default command line options.
- cfgs/
- aws.cfg - connection settings and path to the info on to AWS servers
- jenkins.cfg - connection settings to Jenkins
- biocore.cfg - setting expected structure to biocore info
- rna-seq.template.json - json template for rna-seq pipelines
Others:
* cutadapt.tool_options.cfg
* cwl.tool_options.cfg
* bowtie2.aligner_options.cfg
* fastqc.tool_options.cfg
* trimmomatic.tool_options.cfg
- images/ -- static images of the different associated processes