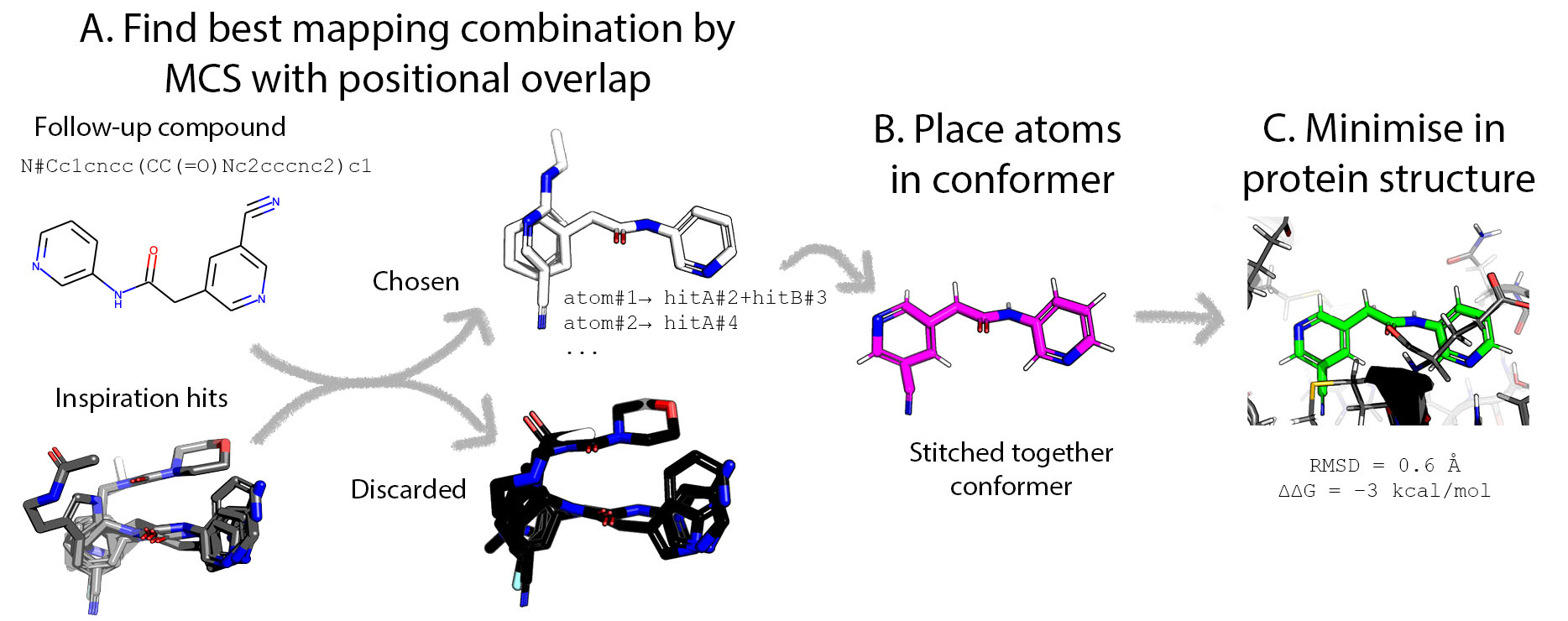
Scaffold hopping between bound compounds by stitching them together like a reanimated corpse.

Given a followup molecule (SMILES) and a series of hits it makes a spatially stitched together version of the followup based on the hits. Like Frankenstein's creation it may violate the laws of chemistry. Planar trigonal topologies may be tetrahedral, bonds unnaturally long etc. This monstrosity is therefore then energy minimised with strong constraints.
Here is an interactive example of mapped molecules.
It is rather tolerant to erroneous/excessive submissions (by automatically excluding them)
and can energy minimise strained conformations.
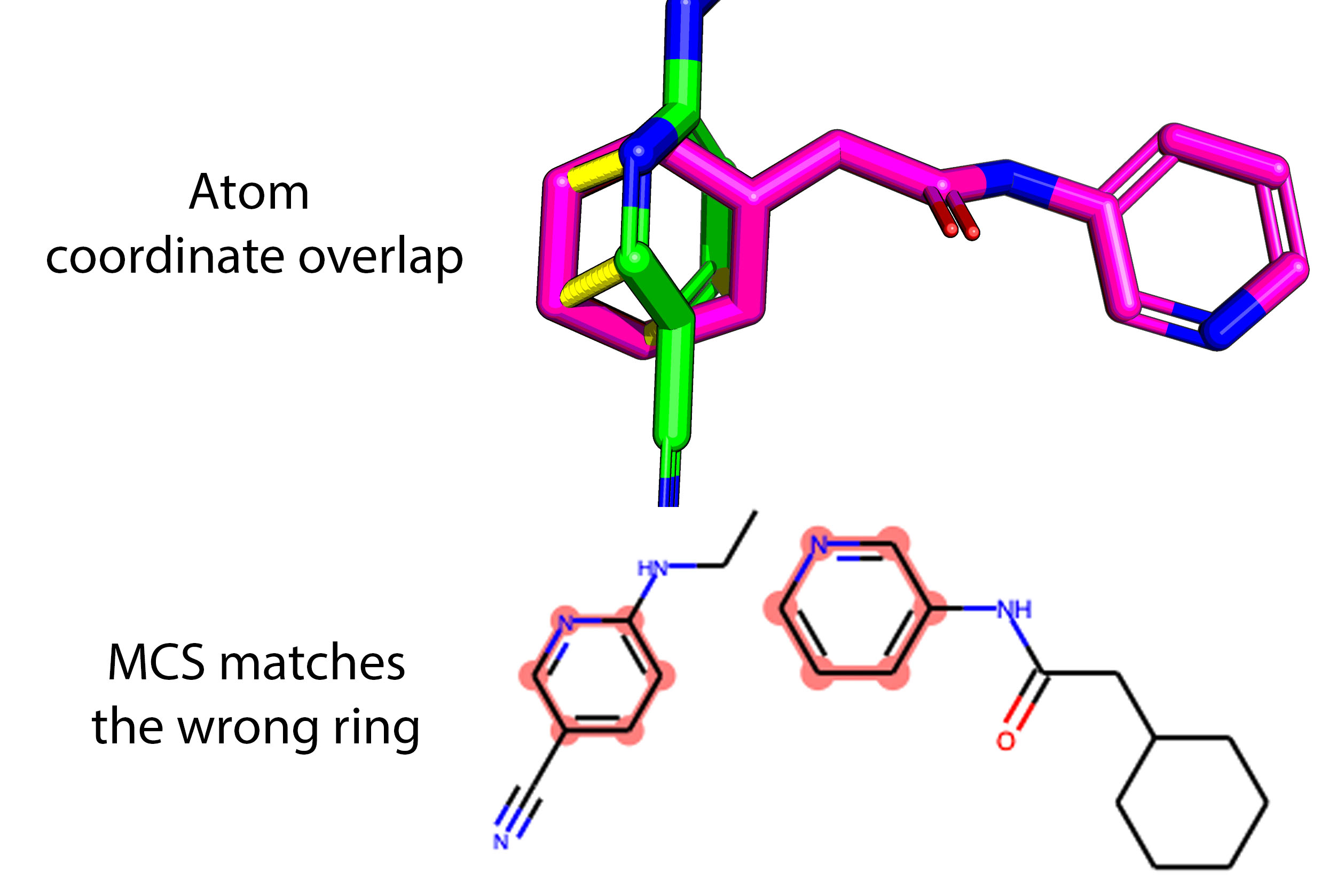
Three mapping approaches were tested, but the key is that hits are pairwise mapped to each other by means of one-to-one atom matching based upon position as opposed to similarity which is easily led astray. For example, note here that the benzene and the pyridine rings overlap, not the two pyridine rings:
It can also merge fragment hits by itself and find the best scoring mergers. It uses the same overlapping position clustering, but also has a decent amount of impossible/uncommon chemistry prevention.
As a consequence, it is not really a docking algorithm as it does not find the pose with the lowest energy within a given volume. Consequently, it is a method to find how faithful is a given followup to the hits provided. Hence the minimised pose should be assessed by the RMSD metric and the ∆∆G score used solely as a cutoff —lower than zero.
Victor, the pipeline, requires my rdkit to params module.
There are three main classes, named after characters from the Fragmenstein book and movies:
Fragmensteinmakes the stitched together molecules — documentationIgoruses PyRosetta to minimise in the protein the fragmenstein followup — documentationVictoris a pipeline that calls the parts, with several features, such as warhead switching —documentation
An honourable mention goes to:
mRMSDis a multiple RMSD variant which does not align and bases which atoms to use on coordinates —documentationrectifieris a class that corrects mistakes in the molecule automatically merged byFragmenstein.
In the absence of pyrosetta (which requires an academic licence), all bar Igor work.
Some changes to the algorithm may happen, see wip.md for more or drop me (matteo) an email.
Fragmenstein was created to see how reasonable are the molecules of fragment mergers submitted in the COVID moonshot project, because after all the underlying method is fragment based screening. This dataset has some unique peculiarities that potentially are not encountered in other projects.
For more see the source code or the Sphinx converted documentation.