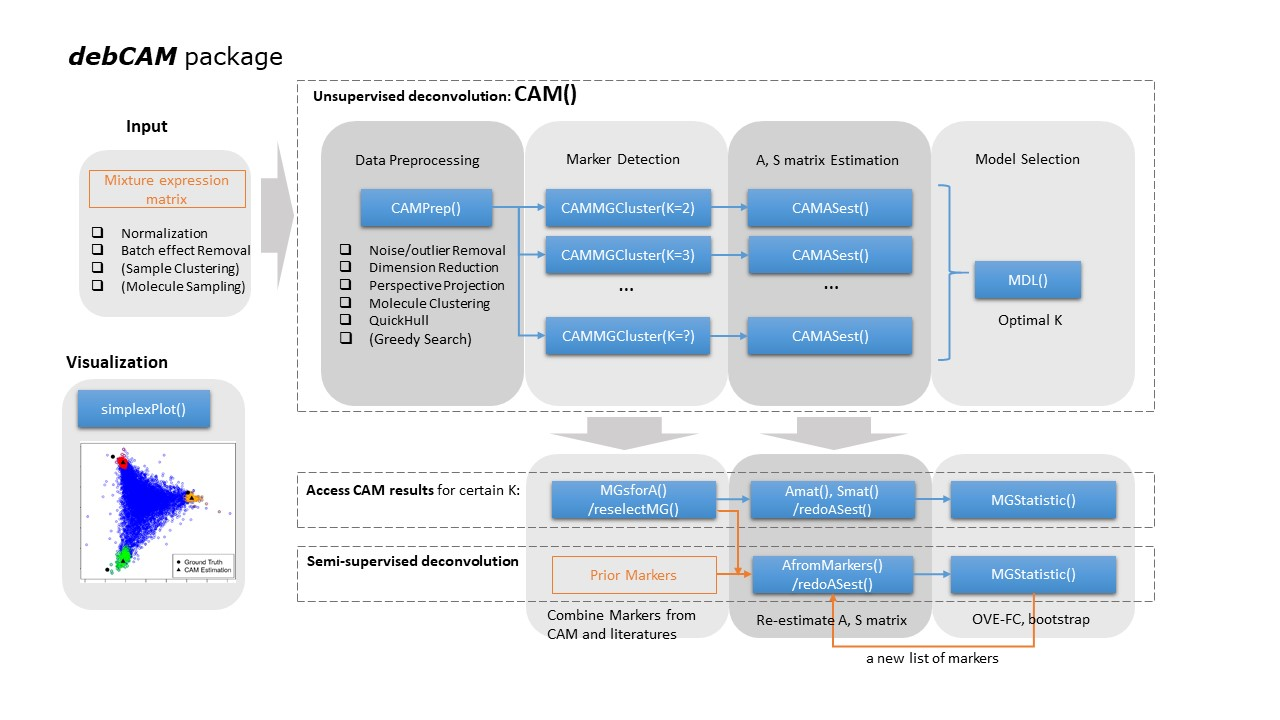
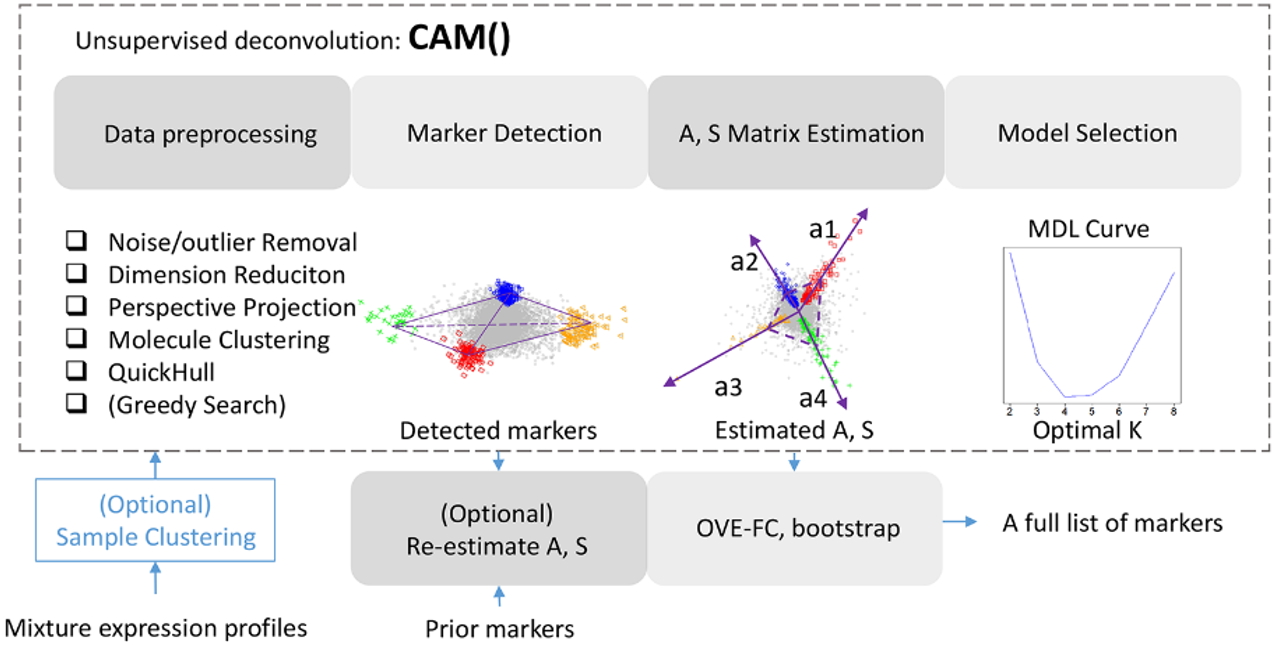
The goal of debCAM is to perform computational deconvolution to dissect complex tissues into molecularly distinctive tissue or cell subtypes based on bulk expression profiles. for tissue heterogeneity characterization. It provides basic functions for unsupervised deconvolution on mixture expression profiles by CAM and some auxiliary functions to help understand the subpopulation-specific results. debCAM also implements functions to perform supervised deconlution based on prior knowledge of molecular markers, S matrix or A matrix. Semi-suprvised deconvolution can also be achieved by combining molecular markers from CAM and from prior knowledge to analyze mixture expressions.
You can install the latest version of debCAM following the instructions in debCAM's bioconductor page: http://bioconductor.org/packages/debCAM.
Or, you can install it from github by
devtools::install_github("Lululuella/debCAM")This is a basic example which shows you how to decompose a data matrix by CAM:
## specify the range of underlying subpopulation number
rCAM <- CAM(data, K = 2:5)The principle underlying CAM is derived from the assumption of well-grounded points among the sources, which can be connected to the concept of molecular markers whose expressions are exclusively enriched in a specific subtype. CAM operates in molecular scatter space, which means CAM does not require the presence of pure samples from each subtype, which is unlikely within complex tissues. Also, only a limited number of subtype-specific markers are needed to guarantee the successful deconvolution by CAM.