The official codes for "Knowledge-enhanced Visual-Language Pretraining for Computational Pathology".
To install Python dependencies:
pip install ./assets/timm_ctp.tar --no-deps
pip install -r requirements.txt
cd ./inference
python easy_inference.py
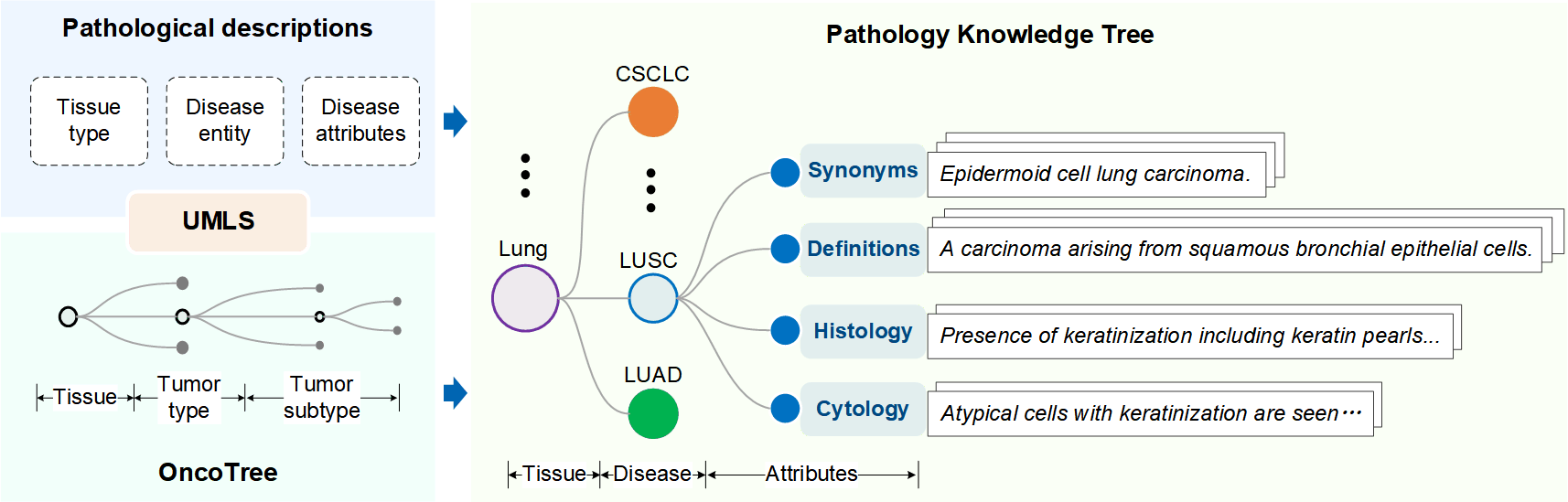
- Pathology Knowledge Tree for knowledge encoding. Based on the structure of OncoTree, we construct a pathology knowledge tree, which contains disease names/synonyms, disease definitions, histology and cytology features.
-
Navigate to OpenPath to download pathology image-text pairs of OpenPath. Note: you might need to download data from Twitter and LAION-5B
-
Navigate to Quilt1m to download pathology image-text pairs of Quilt1m.
-
Retrieval: ARCH.
-
Zero-shot Patch Classification: BACH, NCT-CRC-HE-100K, KatherColon, LC25000, RenalCell, SICAP, SkinCancer, WSSS4LUAD.
-
Zero-shot WSI Tumor Subtyping: TCGA-BRCA, NSCLC: TCGA-LUAD and TCGA-LUSC, TCGA-RCC. Note: The WSIs of TCGA-RCC used in this study keeps the same as MI-Zero
-
CLIP-based: CLIP, BiomedCLIP, PMC-CLIP.
-
SSL-based: CTransPath, PathSSL
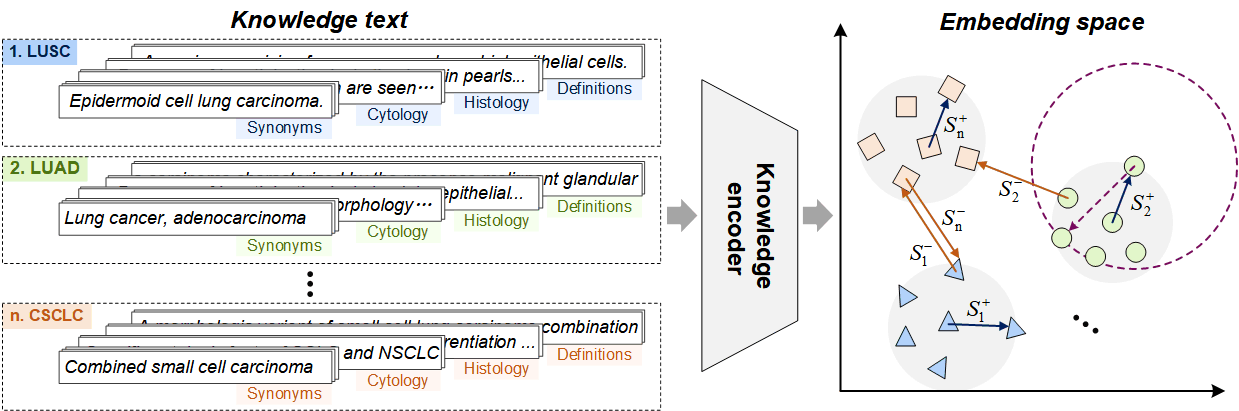
1. Pathology Knowledge Encoding
Projecting tree-structure pathological knowledge into a latent embedding space, where the synonyms, definitions, and corresponding histological/cytological features of the same disease are pulled together while those of different diseases are pushed apart.
Run the following command to perform Pathology Knowledge Encoding:
cd ./S1_knowledge_encoding
python main.py
The main hyper-parameters are summarized in ./S1_knowledge_encoding/bert_training/params.py
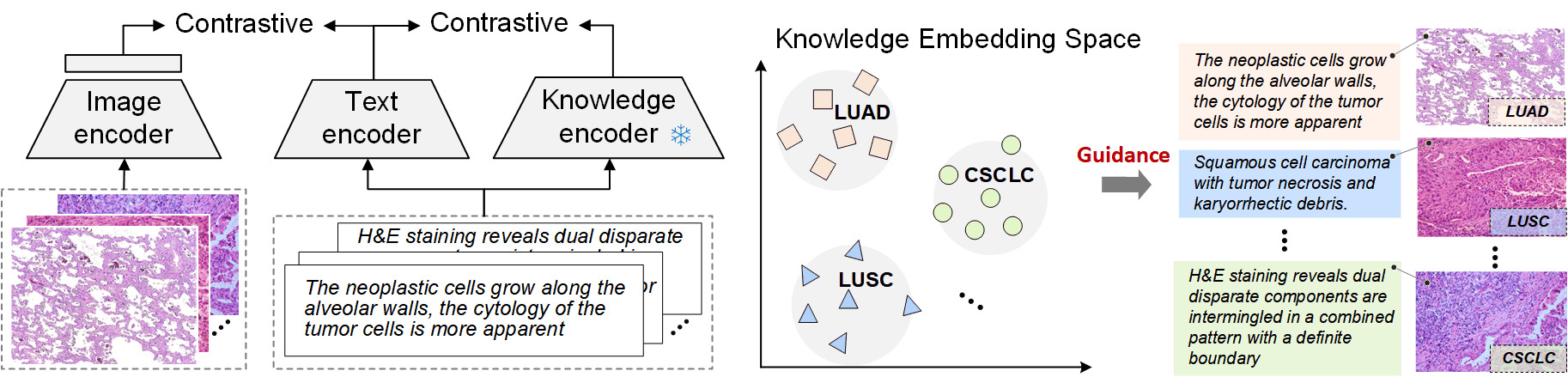
2. Pathology Knowledge Enhanced Pretraining
Leverage the established knowledge encoder to guide visual-language pretraining for computational pathology.
Run the following command to perform Pathology Knowledge Enhanced Pretraining:
cd ./S2_knowledge_enhanced_pretraining
python -m path_training.main
The main hyper-parameters of each model are summarized in ./S1_knowledge_encoding/configs
1. Zero-shot Patch Classification and Cross-modal Retrieval
Run the following command to perform Cross-Modal Retrieval:
cd ./S3_model_eval
python main.py
2. Zero-shot WSI Tumor Subtyping
The code for Zero-shot WSI Tumor Subtyping, please refer to MI-Zero. Note: You might first need to segment WSI into patches by using CLAM, and then implement zeroshot classification on each patch-level images by using MI-Zero.
We provide the models' checkpoints for KEP-32_OpenPath, KEP-16_OpenPath, KEP-CTP_OpenPath, KEP-32_Quilt1m, KEP-16_Quilt1m, KEP-CTP_Quilt1m, which can be download from BaiduPan or from google drive with link: GoogleDrive.
If you use this code for your research or project, please cite:
@misc{zhou2024knowledgeenhanced,
title={Knowledge-enhanced Visual-Language Pretraining for Computational Pathology},
author={Xiao Zhou and Xiaoman Zhang and Chaoyi Wu and Ya Zhang and Weidi Xie and Yanfeng Wang},
year={2024},
eprint={2404.09942},
archivePrefix={arXiv},
primaryClass={cs.CV}
}