- 3D RNA-seq App v2.0.1 (15/08/2020).
- Previous versions can be found in https://github.com/wyguo/ThreeDRNAseq/releases
3D RNA-seq is currently under a dual-licensing model.
- Open source under GPLV3.0. For academic and non-commercial use, it is free.
- Commercial. For commercial use, please get in touch to obtain commercial licenses. Contact us
- Description
- User group
- Prior to 3D RNA-seq
- Capability of 3D RNA-seq
- Run 3D RNA-seq App
- How to get help
- Pipeline of 3D RNA-seq App
- References
To see a demo video, click the screenshot

The ThreeDRNAseq (3D RNA-seq) R package provides an interactive graphical user interface (GUI) for RNA-seq data analysis using accurate quantification of RNA-seq reads. It allows users to perform differential expression (DE), differential alternative splicing (DAS) and differential transcript usage (DTU) (3D) analyses based on a popular pipeline limma (Ritchie et al., 2015). The 3D RNA-seq GUI is based on R shiny App and enables a complete RNA-seq analysis to be done within only 3 Days (3D). To use our pipeline in your work, please cite:
- Guo,W., Tzioutziou,N., Stephen,G., Milne,I., Calixto,C., Waugh,R., Brown,J.W., and Zhang,R. (2019) 3D RNA-seq - a powerful and flexible tool for rapid and accurate differential expression and alternative splicing analysis of RNA-seq data for biologists. bioRxiv, 656686. doi: https://doi.org/10.1101/656686.
- Calixto,C.P.G., Guo,W., James,A.B., Tzioutziou,N.A., Entizne,J.C., Panter,P.E., Knight,H., Nimmo,H.G., Zhang,R., and Brown,J.W.S. (2018) Rapid and Dynamic Alternative Splicing Impacts the Arabidopsis Cold Response Transcriptome. Plant Cell, 30, 1424–1444.
To report bugs, leave comments, ask questions and post discussions, please go to:
- Group group: https://groups.google.com/forum/#!forum/3d-rna-seq-app-user-group
- Or Github Issues: https://github.com/wyguo/ThreeDRNAseq/issues
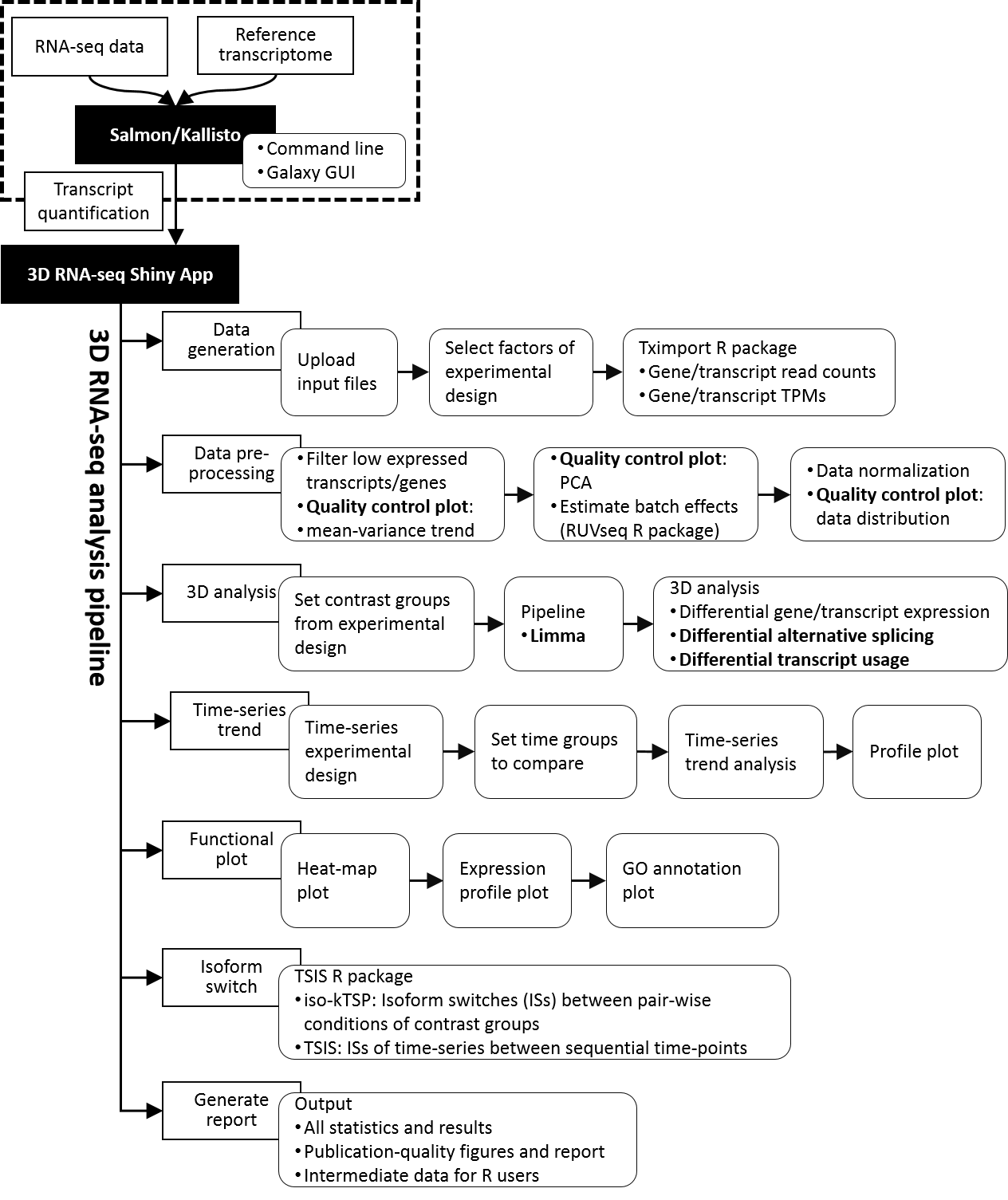
RNA-seq raw read data is organised by sample. Different samples are either generated by different experimental conditions (multiple genotypes, tissue types or treatments, time-of day circadian experiments) or by biological replication (sometimes also sequencing replication) and each of them is used for the construction of individual libraries. Next, quality control and trimming of the RNA-seq reads should be performed to ensure that high quality RNA-seq reads are used for downstream analysis. The read data are then used for RNA-seq quantification. This step can be performed using many different pipelines, and the type of pipeline determines whether you can use 3D RNA-seq for your downstream expression analyses or not. 3D RNA-seq is only compatible with transcript quantification data derived from Salmon (Patro et al., 2017) or Kallisto (Bray et al., 2016) with the use of a reference transcriptome or Reference Transcript Dataset which contains a list of the known genes and transcripts for the organism under study. The Salmon/Kallisto output file contains the TPM values for each transcript organised by biological repeat and treatment(s). Depending on the size of the dataset, the transcript quantification procedure might take up to 1-2 days.
The 3D RNA-seq analysis pipeline starts with a number of steps to pre-process the data and reduce noise (e.g. low expression filters based on expression mean-variance trend, visualise data variation, batch effect estimation, data normalisation, etc.). The optimal parameters for each step are determined by quality control plots (e.g. mean-variance trend plots, PCA plots, data distribution plots, etc.). Stringent filters are applied to statistical testing of 3D genes/transcripts to control false positives. After the analysis, publication quality plots (e.g. expression profile plots, heatmap, GO annotation plots, etc.) and reports can be generated. The entire 3D RNA-seq analysis takes only 1 Day or less and all actions are performed by simple mouse clicks on the App.
User manual: https://github.com/wyguo/ThreeDRNAseq/blob/master/vignettes/user_manuals/3D_RNA-seq_App_manual.md
The 3D RNA-seq App docker image is hosted by the James Hutton Institute server. Open the App by this link: https://ics.hutton.ac.uk/3drnaseq
To perform 3D analysis, no installation is required. Users need to upload input data to our server. All results, reports, figures and intermediate data will be zipped and downloaded in the final step.
To run the 3D RNA-seq App through RStudio on a local PC, users do not need to upload the data to our server and all the outputs will be directly saved to the App working directory. Please run the following command to install ThreeDRNAseq R package and the packages of dependencies.
Install dependency packages
R version required > 4.0.0
#####################################################################################
## Install packages of dependency
###---> Install packages from Cran
cran.package.list <- c("shiny","shinydashboard","rhandsontable","shinyFiles",
"shinyjs","shinyBS","shinyhelper","shinyWidgets",
"magrittr","DT","plotly","ggplot2","eulerr",
"gridExtra","grid","fastcluster","rmarkdown","base64enc",
"ggrepel","zoo","gtools")
for(i in cran.package.list){
if(!(i %in% rownames(installed.packages()))){
message('Installing package: ',i)
install.packages(i,dependencies = T)
} else next
}
###---> Install packages from Bioconductor
bioconductor.package.list <- c('tximport','edgeR','limma','RUVSeq',
'ComplexHeatmap','rhdf5')
for(i in bioconductor.package.list){
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
if(!(i %in% rownames(installed.packages()))){
message('Installing package: ',i)
BiocManager::install(i,dependencies = T)
} else next
}Install ThreeDRNAseq R package
ThreeDRNAseq R package can be installed from Github by using devtools R package
##################################################################################################
## use devtools R package to install ThreeDRNAseq from Github
###---> If devtools is not installed, please install
if(!requireNamespace("devtools", quietly = TRUE))
install.packages('devtools',dependencies = TRUE)
###---> Install ThreeDRNAseq
if(!requireNamespace("ThreeDRNAseq", quietly = TRUE))
devtools::install_github('wyguo/ThreeDRNAseq')Run 3D RNA-seq App
library(ThreeDRNAseq)
run3DApp()The ThreeDRNAseq R package can be used as a normal R package. The vignette of command line for 3D analysis can be found in:
https://github.com/wyguo/ThreeDRNAseq/blob/master/vignettes/user_manuals/3D_RNA-seq_command_line_user_manual.md
NOTE:
- If you are working on RNA-seq data with very big size of transcript quantification (≥ 2GB), it is recommended to remove the redundant files in the Salmon/Kallisto outputs (see Input files) to reduce data size or run the 3D RNA-seq App through RStudio on a local PC.
- To run the ThreeDRNAseq App on a local PC, if any other denpendency R packages are missing, please install them.
3D RNA-seq App "easy-to-use" manual and command line manual:
https://github.com/wyguo/ThreeDRNAseq/blob/master/vignettes/user_manuals
Tooltips:
In the GUI, users can click tooltips in specific steps for help information.
Contact us:
3D RNA-seq App is developed and maintained by Dr. Wenbin Guo from Plant Sciences Division, School of Life Sciences, University of Dundee. If you have any questions and suggestions, please contact:
- Dr. Wenbin Guo: wenbin.guo@hutton.ac.uk
- Dr. Runxuan Zhang: runxuan.zhang@hutton.ac.uk
- Bray,N.L., Pimentel,H., Melsted,P., and Pachter,L. (2016) Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol., 34, 525–527.
- Calixto,C.P.G., Guo,W., James,A.B., Tzioutziou,N.A., Entizne,J.C., Panter,P.E., Knight,H., Nimmo,H.G., Zhang,R., and Brown,J.W.S. (2018) Rapid and Dynamic Alternative Splicing Impacts the Arabidopsis Cold Response Transcriptome. Plant Cell, 30, 1424–1444.
- Guo,W., Tzioutziou,N., Stephen,G., Milne,I., Calixto,C., Waugh,R., Brown,J.W., and Zhang,R. (2019) 3D RNA-seq - a powerful and flexible tool for rapid and accurate differential expression and alternative splicing analysis of RNA-seq data for biologists. bioRxiv, 656686. doi: https://doi.org/10.1101/656686.
- Patro,R., Duggal,G., Love,M.I., Irizarry,R.A., and Kingsford,C. (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods, 14, 417–419.
- Ritchie,M.E., Phipson,B., Wu,D., Hu,Y., Law,C.W., Shi,W., and Smyth,G.K. (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res, 43, e47.
- Robinson,M.D., McCarthy,D.J., and Smyth,G.K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–40.