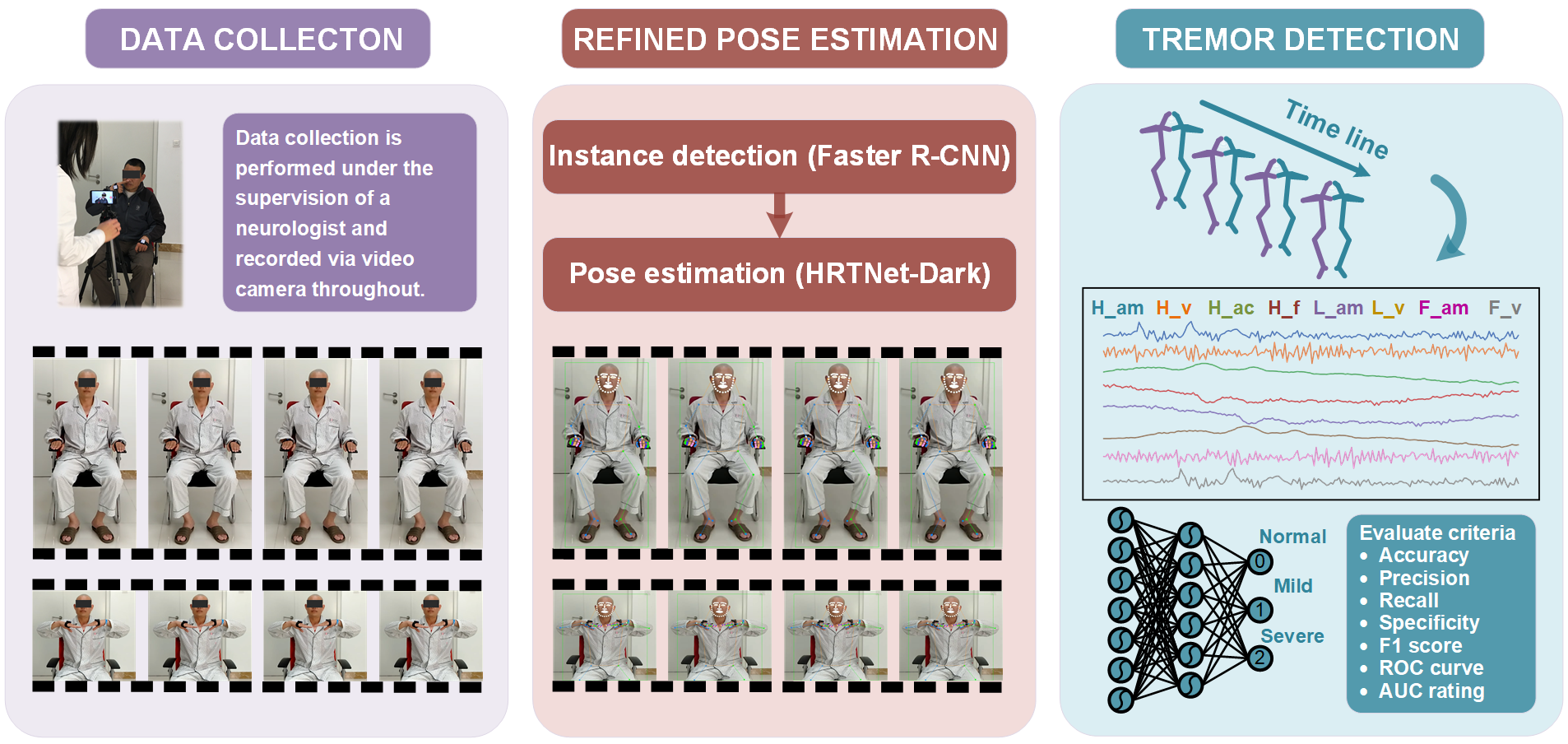
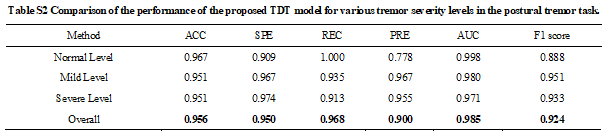
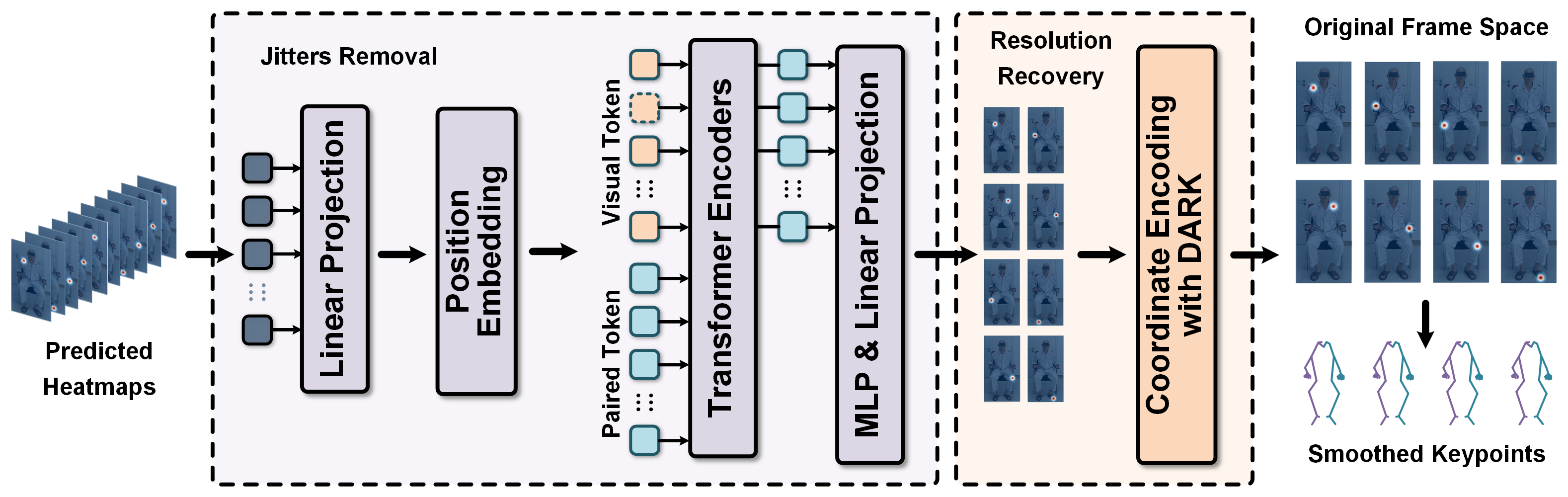
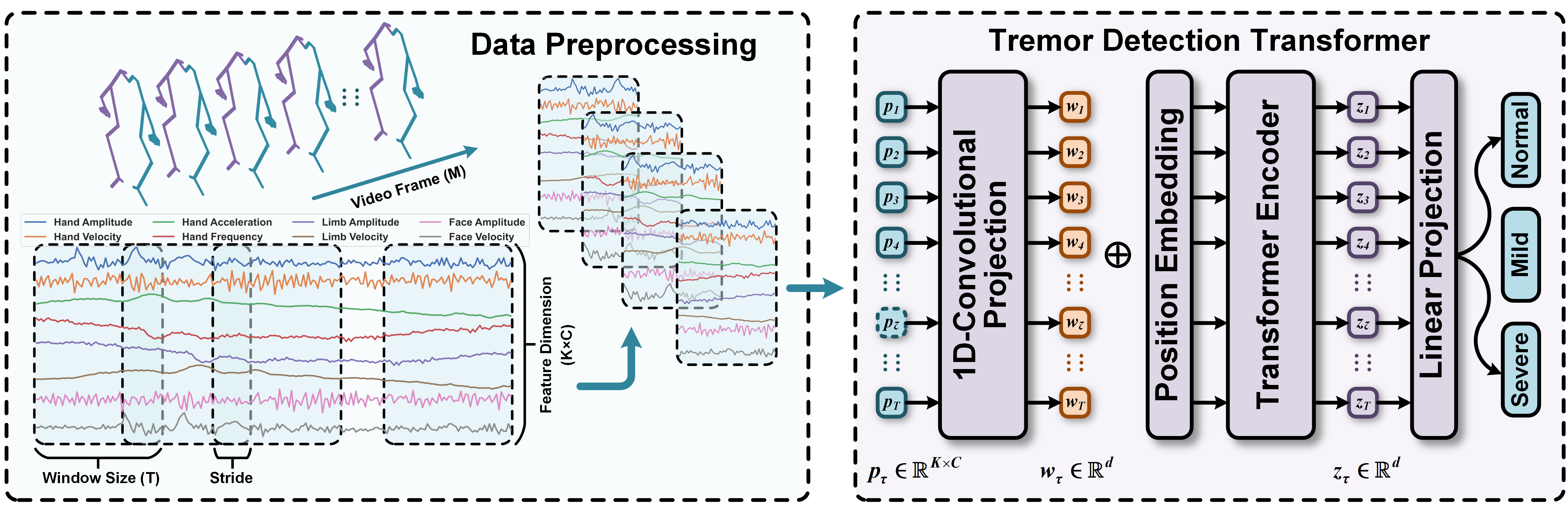
Tremor Detection Transformer: An Automatic Symptom Assessment Framework Based on Refined Whole-Body Pose Estimation
We used public COCO Whole-body dataset and our collected private PLAGH dataset in this study.
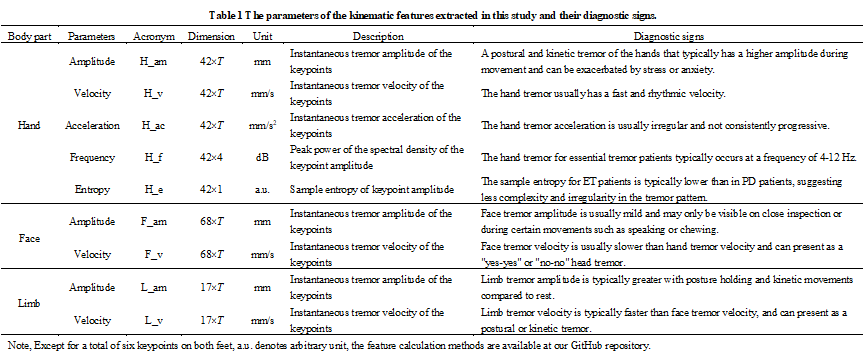
Table 1 details each kinematic parameters, acronyms, dimensions, unit, description, and diagnostic signs.
Detailed feature calculation methods
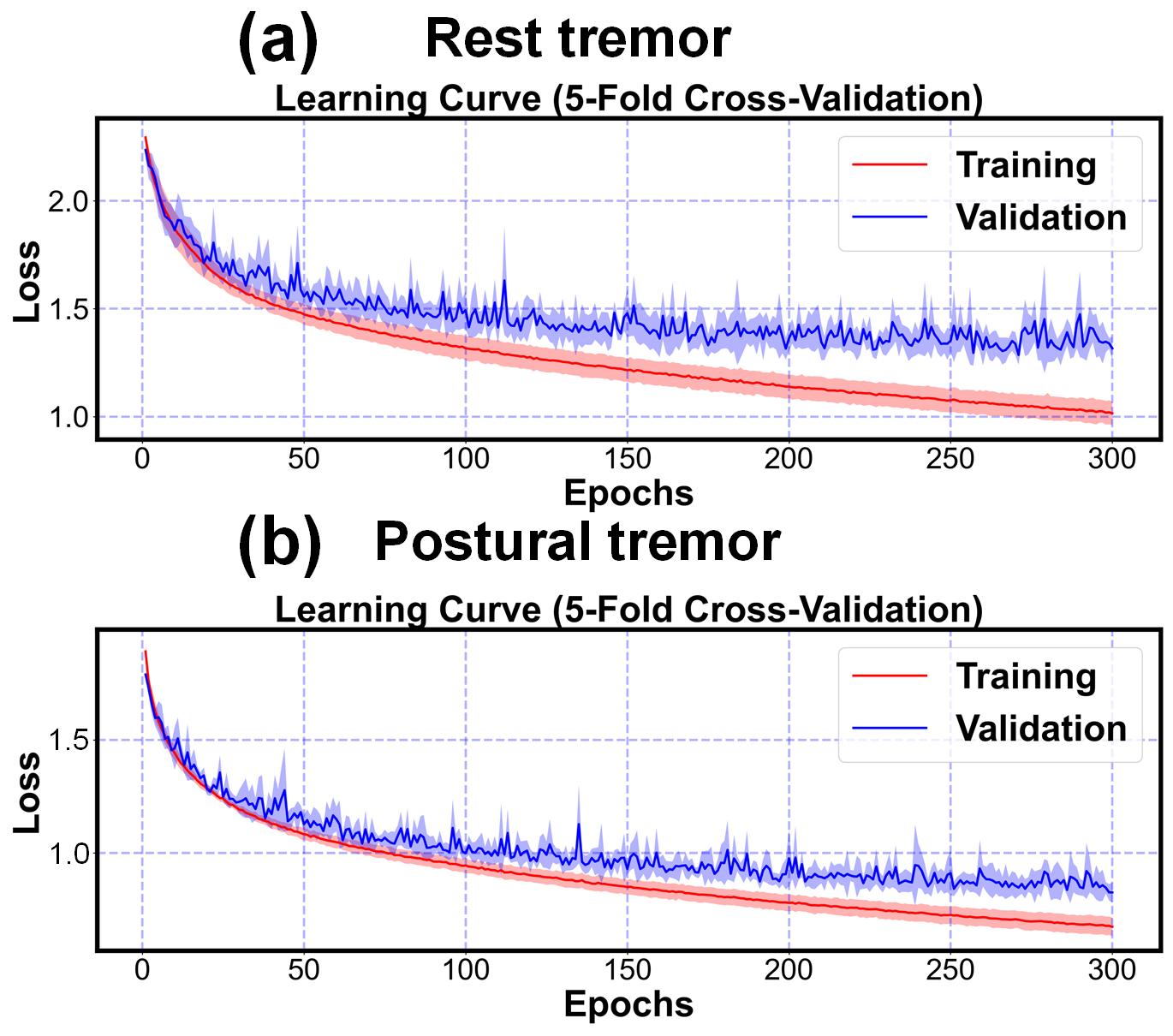
To evaluate the model fitting performance, we present the learning curves of our proposed TDT model for both rest tremor and postural tremor tasks in Fig. S1 in the supplementary material, respectively, which correspond to the results of the five-fold cross-validation experiments. The solid segment of each curve reflects the average loss, while the shaded segment represents the standard deviation of the five validations. These results affirm the robust learning ability of the TDT model.
Fig. S1. Learning curves of the proposed TDT model in the rest and postural tremor tasks, respectively. The solid line indicates the mean of the five-fold cross-validation and the shaded part is the standard deviation.
- Python3
- Pytorch==1.7
- Numpy==1.20.1
- scikit-learn==0.24.1
- Pandas==1.2.4
- skorch==0.10.0 (For DEV risk calculations)
- openpyxl==3.0.7 (for classification reports)
- Wandb=0.12.7 (for sweeps)
To add new dataset (e.g., NewData), it should be placed in a folder named: NewData in the datasets directory.
Since "NewData" has several domains, each domain should be split into train/test splits with naming style as "train_i_x.pt" and "test_i_x.pt" for each i-th fold.
The structure of data files should in dictionary form as follows:
train.pt = {"samples": data, "labels: labels}, and similarly for test.pt.
Next, you have to add a class with the name NewData in the configs/data_model_configs.py file.
You can find similar classes for existing datasets as guidelines.
Also, you have to specify the cross-domain scenarios in self.scenarios variable.
Last, you have to add another class with the name NewData in the configs/hparams.py file to specify
the training parameters.
To add a new algorithm, place it in algorithms/algorithms.py file.
The experiments are organised in a hierarchical way such that:
- Several experiments are collected under one directory assigned by
--experiment_description. - Each experiment could have different trials, each is specified by
--run_description. - For example, if we want to experiment different Tremor Detection methods with CNN backbone, we can assign
--experiment_description CNN_backnones --run_description TDModeland--experiment_description CNN_backnones --run_description TDTand so on.
To train a model:
python main.py --experiment_description exp1 \
--run_description run_1 \
--da_method DANN \
--dataset HHAR \
--backbone CNN \
--num_runs 5 \
--is_sweep False
Sweeps here are deployed on Wandb, which makes it easier for visualization, following the training progress, organizing sweeps, and collecting results.
python main.py --experiment_description exp1_sweep \
--run_description sweep_over_lr \
--da_method TCN \
--dataset PLAGH \
--backbone CNN \
--num_runs 200 \
--is_sweep True \
--num_sweeps 50 \
--sweep_project_wandb TEST
Upon the run, you will find the running progress in the specified project page in wandb.
Note: If you got cuda out of memory error during testing, this is probably due to DEV risk calculations.
To obtain the source-only or the target-only results, you can run same_domain_trainer.py file.
- Each run will have all the cross-domain scenarios results in the format
src_to_trg_run_x, wherexis the run_id (you can have multiple runs by assigning--num_runsarg). - Under each directory, you will find the classification report, a log file, checkpoint, and the different risks scores.
- By the end of the all the runs, you will find the overall average and std results in the run directory.
If you found this work useful for you, please consider citing it.
@article{TDT,
title = {Tremor Detection Transformer: An Automatic Symptom Assessment Framework Based on Refined Whole-Body Pose Estimation},
author = {Chenbin Ma, Lishuang Guo, Longsheng Pan, Xuemei Li, Chunyu Yin, Rui Zong, Zhengbo Zhang},
journal = {####},
year = {2023}
}
For any issues/questions regarding the paper or reproducing the results, please contact any of the following.
Chenbin Ma: machenbin@buaa.edu.cn
Zhengbo Zhang: zhengbozhang@126.com
Department of Biomedical Engineering, Chinese PLA General Hospital, 28 Fuxing Road, Beijing, 100853