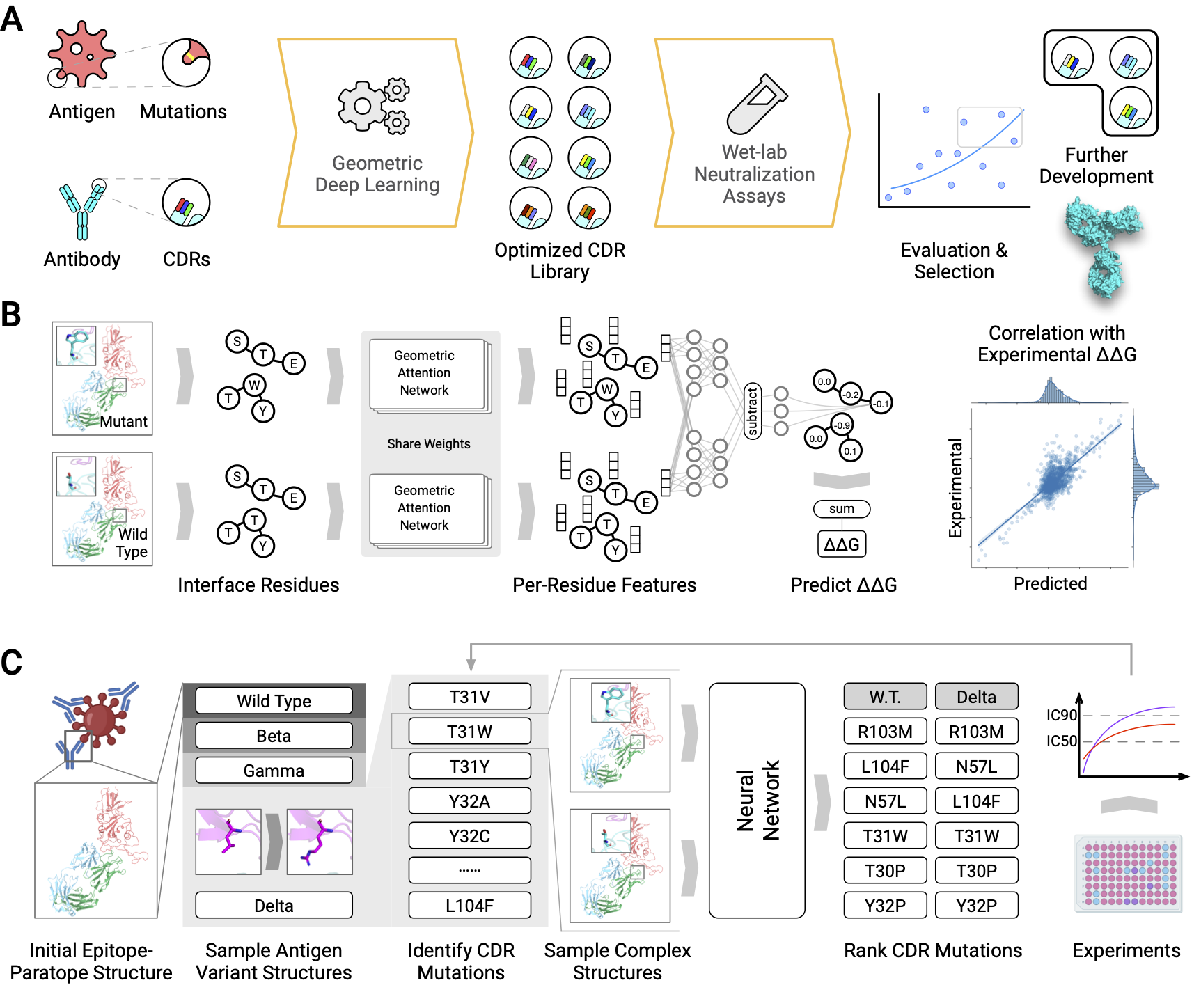
This repository contains the deep learning model introduced in the paper "Deep Learning-Guided Optimization of Human Antibody Against SARS-CoV-2 Variants with Broad Neutralization". It predicts changes in binding energy upon mutation (ddG) for protein-protein complexes.
- Please check out our latest work on mutational effect prediction for protein-protein interactions: Rotamer Density Estimator is an Unsupervised Learner of the Effect of Mutations on Protein-Protein Interaction (ICLR 2023) [Code] [Paper]
The model is tested with Python 3.8, PyTorch 1.10, cudatoolkit 11.3, and Biopython 1.79. The dependencies can be set up using the following commands:
conda create --name ddg-predict python=3.8 -y
conda activate ddg-predict
conda install pytorch=1.10.2 cudatoolkit=11.3 -c pytorch -y
conda install biopython=1.79 easydict -c conda-forge -yNext, clone this repository by:
git clone https://github.com/HeliXonProtein/binding-ddg-predictor.git
cd binding-ddg-predictorThe model requires two input PDB files: (1) a wild-type complex structure, and (2) a mutated complex structure. The mutated structures are typically built by protein design packages such as Rosetta. Note that both structures must have the same length. The ddG can be predicted for the two structures by running the command:
python ./scripts/predict.py <path-to-wild-type-pdb> <path-to-mutant-pdb>A quick example can be obtained by running:
python ./scripts/predict.py ./data/example_wt.pdb ./data/example_mut.pdb
Coming soon...
Please contact luost[at]helixon.com for any questions related to the source code.