This project is an implementation of Multimodal Model-Agnostic Meta-Learning via Task-Aware Modulation, which is published in NeurIPS 2019. Please visit our project page for more information and contact Hexiang Hu for any questions.
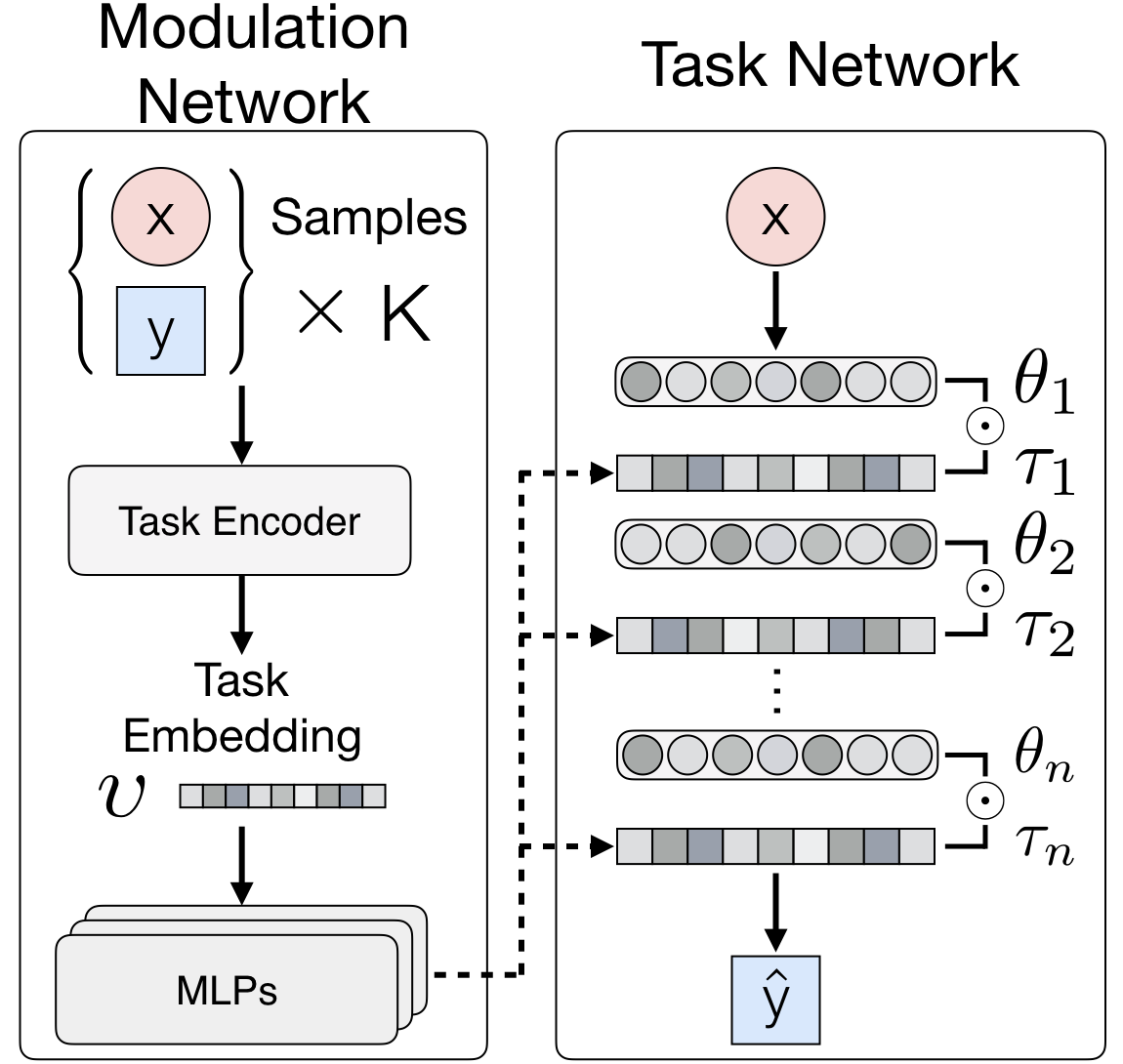
Model-agnostic meta-learners aim to acquire meta-prior parameters from a distribution of tasks and adapt to novel tasks with few gradient updates. Yet, seeking a common initialization shared across the entire task distribution substantially limits the diversity of the task distributions that they are able to learn from. We propose a multimodal MAML (MMAML) framework, which is able to modulate its meta-learned prior according to the identified mode, allowing more efficient fast adaptation. An illustration of the proposed framework is as follows.
Use of Conda Environment is suggested to for straightforward handling of the dependencies.
conda env create -f environment.yml
conda activate mmaml_regressionAfter installation, we can start to train models with the following commands.
python main.py --dataset mixed --num-batches 70000 --model-type fc --fast-lr 0.001 --meta-batch-size 50 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 5 --output-folder 2mods-maml-5steps --bias-transformation-size 20 --disable-norm
python main.py --dataset mixed --num-batches 70000 --model-type multi --fast-lr 0.001 --meta-batch-size 50 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 5 --output-folder 2mods-multi-maml-5steps --bias-transformation-size 20 --disable-norm
python main.py --dataset mixed --num-batches 70000 --model-type gated --fast-lr 0.001 --meta-batch-size 50 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 5 --output-folder 2mods-mmaml-5steps --bias-transformation-size 20 --disable-norm --embedding-type LSTM --embedding-dims 200 --inner-loop-grad-clip 10
python main.py --dataset mixed --num-batches 70000 --model-type gated --fast-lr 0.001 --meta-batch-size 50 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 5 --output-folder 2mods-mmaml-sigmoid-5steps --bias-transformation-size 20 --disable-norm --embedding-type LSTM --embedding-dims 100 --inner-loop-grad-clip 10 --condition-type sigmoid_gate
python main.py --dataset mixed --num-batches 70000 --model-type gated --fast-lr 0.001 --meta-batch-size 50 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 5 --output-folder 2mods-mmaml-softmax-5steps --bias-transformation-size 20 --disable-norm --embedding-type LSTM --embedding-dims 100 --inner-loop-grad-clip 10 --condition-type softmax
python main.py --dataset mixed --num-batches 70000 --model-type gated --fast-lr 0.0 --meta-batch-size 50 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 1 --output-folder 2mods-mmaml-pre-1steps --bias-transformation-size 20 --disable-norm --embedding-type LSTM --embedding-dims 100 --inner-loop-grad-clip 10
python main.py --dataset many --num-batches 70000 --model-type fc --fast-lr 0.001 --meta-batch-size 75 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 5 --output-folder 3mods-maml-5steps --bias-transformation-size 20 --disable-norm
python main.py --dataset many --num-batches 70000 --model-type multi --fast-lr 0.001 --meta-batch-size 75 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 5 --output-folder 3mods-multi-maml-5steps --bias-transformation-size 20 --disable-norm
python main.py --dataset many --num-batches 70000 --model-type gated --fast-lr 0.001 --meta-batch-size 75 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 5 --output-folder 3mods-mmaml-5steps --bias-transformation-size 20 --disable-norm --embedding-type LSTM --embedding-dims 200 200 200 --inner-loop-grad-clip 10
python main.py --dataset many --num-batches 70000 --model-type gated --fast-lr 0.001 --meta-batch-size 75 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 5 --output-folder 3mods-mmaml-sigmoid-5steps --bias-transformation-size 20 --disable-norm --embedding-type LSTM --embedding-dims 100 100 100 --inner-loop-grad-clip 10 --condition-type sigmoid_gate
python main.py --dataset many --num-batches 70000 --model-type gated --fast-lr 0.001 --meta-batch-size 75 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 5 --output-folder 3mods-mmaml-softmax-5steps --bias-transformation-size 20 --disable-norm --embedding-type LSTM --embedding-dims 100 100 100 --inner-loop-grad-clip 10 --condition-type softmax
python main.py --dataset many --num-batches 70000 --model-type gated --fast-lr 0.00 --meta-batch-size 75 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 1 --output-folder 3mods-mmaml-pre-5steps --bias-transformation-size 20 --disable-norm --embedding-type LSTM --embedding-dims 200 200 200 --inner-loop-grad-clip 10
python main.py --dataset five --num-batches 70000 --model-type fc --fast-lr 0.001 --meta-batch-size 125 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 5 --output-folder 5mods-maml-5steps --bias-transformation-size 20 --disable-norm
python main.py --dataset five --num-batches 70000 --model-type multi --fast-lr 0.001 --meta-batch-size 125 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 5 --output-folder 5mods-multi-maml-5steps --bias-transformation-size 20 --disable-norm
python main.py --dataset five --num-batches 70000 --model-type gated --fast-lr 0.001 --meta-batch-size 125 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 5 --output-folder 5mods-mmaml-5steps --bias-transformation-size 20 --disable-norm --embedding-type LSTM --embedding-dims 200 200 200 --inner-loop-grad-clip 10
python main.py --dataset five --num-batches 70000 --model-type gated --fast-lr 0.001 --meta-batch-size 125 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 5 --output-folder 3mods-mmaml-sigmoid-5steps --bias-transformation-size 20 --disable-norm --embedding-type LSTM --embedding-dims 100 100 100 --inner-loop-grad-clip 10 --condition-type sigmoid_gate
python main.py --dataset five --num-batches 70000 --model-type gated --fast-lr 0.001 --meta-batch-size 125 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 5 --output-folder 3mods-mmaml-softmax-5steps --bias-transformation-size 20 --disable-norm --embedding-type LSTM --embedding-dims 100 100 100 --inner-loop-grad-clip 10 --condition-type softmax
python main.py --dataset five --num-batches 70000 --model-type gated --fast-lr 0.00 --meta-batch-size 125 --num-samples-per-class 10 --num-val-samples 5 --noise-std 0.3 --hidden-sizes 100 100 100 --device cuda --num-updates 1 --output-folder 5mods-mmaml-pre-5steps --bias-transformation-size 20 --disable-norm --embedding-type LSTM --embedding-dims 200 200 200 --inner-loop-grad-clip 10