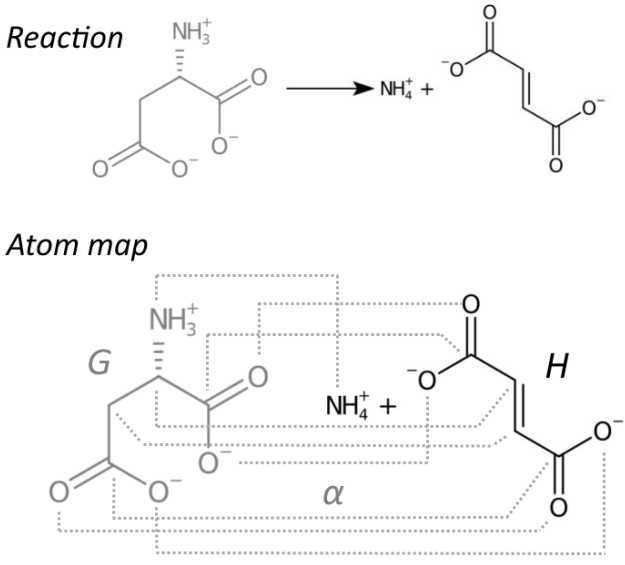
Evaluation of the Equivalence of Atom-to-Atom Maps
Center for Scalable Data Analytics and Artificial Intelligence, Leipzig / Dresden, Germany. See ScaDS.AI.
Bioinformatics Group, Department of Computer Science, Leipzig University, Germany. See Bioinf.
Interdisciplinary Center for Bioinformatics, Leipzig University, Germany. See IZBI.
-
Marcos E. González Laffitte
Bioinformatics Group and ScaDS.AI, Leipzig University
marcoslaffitte@gmail.com
marcos@bioinf.uni-leipzig.de -
Nora Beier
Bioinformatics Group, Leipzig University
nora@bioinf.uni-leipzig.de -
Nico Domschke
Bioinformatics Group, Leipzig University
dnico@bioinf.uni-leipzig.de -
Prof. Dr. Peter F. Stadler
Bioinformatics Group and ScaDS.AI and Interdisciplinary Center for Bioinformatics, Leipzig University
studla@bioinf.uni-leipzig.de
Note: the atom maps to be compared need to be complete, i.e., bijective. An incomplete atom map is that in which there are atoms on one side of the reaction not being mapped to atoms on the other side, in other words, the atom map does not represent a bijection. This in turn requires for the analyzed reactions to be stoichiometrically balanced. For more information see the following reference.
This repository was developed as part of the contribution:
[1] M. E. González Laffitte, N. Beier, N. Domschke, P. F. Stadler, Comparison of Atom Maps. MATCH Commun. Math. Comput. Chem. 90 (2023) 75–102.
Link: https://match.pmf.kg.ac.rs/issues/m90n1/m90n1_75-102.html
In order to run these programs you will require some python packages, below we show how to install them directly into an anaconda environment. In particular, if you are runnig MappingTool you will need to install Java or OpenJDK in your computer (required to run the RDT mapper). After this you only need a list of unannotated reaction SMILES inside a plain-text file, over which you can apply the pipeline NumberingTool > MappingTool > EEquAAM, meaning that the output of one program is the input for the following. Below you can find the python commands to run each program.
conda create -n eequaam python=3.9.13 networkx=2.8.4 matplotlib=3.5.2 numpy=1.21.5 beautifulsoup4=4.11.1
conda activate eequaam
pip install pysmiles==1.0.2 rdkit==2022.9.3 graphkit-learn==0.2.1 rxnmapper==0.2.4 chytorch-rxnmap==1.3 chython==1.54 chytorch==1.27 torch==1.11.0 && pip install --no-warn-conflicts transformers==4.24.0
Remember to always activate the eequaam conda environment before using the programs in this repository.
If your initial list of unannotated reaction SMILES doesn't have identifiers for each reaction, you can make use of this script to add them to your list. In the following command [myFile.smiles] is a plain-text file whose lines are each a reaction SMILES. Please mind the *.smiles extension:
python NumberingTool.py [myFile.smiles]
The output will be a plain-text file [myFile_id.smiles] whose lines alternate between the reaction SMILES and their identifiers. See NumberingTool for an example of the input and output formats.
If you already have a list of unannotated reaction SMILES with identifiers and you want to produce atom maps for these reactions, you can make use of this script. This wraps the three atom mapping tools RXN, RDT and Graphormer.
python MappingTool.py [myFile_id.smiles]
The output will include a plain-text file [myFile_aam.smiles] containing the annotated SMILES of those suitable and balanced reactions that were completely mapped by the three mappers. The extension of these files is again *.smiles. Nevertheless they have different formats. See MappingTool for an example of the input and output formats.
There are two options when running EEquAAM. The first evaluates the equivalence of atom maps only through the ITS method. This is the fastest method and therefore the default option:
python EEquAAM.py [myFile_aam.smiles]
The second option evaluates the equivalence of atom maps with the three methods ITS, AUX and ISO described in [1]. These are mathematically equivalent, but the ISO method may be more time consuming depending on the number of symmetries of the involved molecules. To run and compare these methods use the following command:
python EEquAAM.py --sanity-check [myFile_aam.smiles]
The output will include a summary indicating for which reactions the given maps were all equivalent, and for which the given maps were non-equivalent. See EEquAAM for an example of the input and output formats.
conda deactivate
conda env remove -n eequaam
-
RXNMapper [RXN]
http://rxnmapper.ai/
https://pypi.org/project/rxnmapper/ -
Reaction Decoder Tool [RDT]
https://academic.oup.com/bioinformatics/article/32/13/2065/1743096
https://github.com/asad/ReactionDecoder
https://github.com/asad/ReactionDecoder/releases -
GraphormerMapper [Graphormer]
https://pubs.acs.org/doi/10.1021/acs.jcim.2c00344
https://github.com/chython/chytorch-rxnmap
https://pypi.org/project/chytorch-rxnmap/ -
General specifications on both anotated and unannotated SMILES
http://opensmiles.org/opensmiles.html
The programs in this repository are part of the work published in
https://match.pmf.kg.ac.rs/issues/m90n1/m90n1_75-102.html
and are released under
MIT License Copyright (c) 2023 Marcos E. González Laffitte
See LICENSE file for full license details.