An R-package for identifying model violations under the Generalized Mixed Yule Coalescent (GMYC) model
P2C2M.GMYC can be installed from Github using the function install_github implemented in devtools R-package.
library(devtools)
devtools::install_github("emanuelmfonseca/P2C2M.GMYC")
P2C2M automatically installs all the dependencies, but if you get errors, try to install them individually. Here is the list of all required packages:
install.packages("ape")
install.packages("pegas")
install.packages("phangorn")
install.packages("phyclust")
install.packages("pegas")
install.packages("TreeSim")
install.packages("splits", repos="http://R-Forge.R-project.org", type="source")
install_github("liamrevell/phytools")
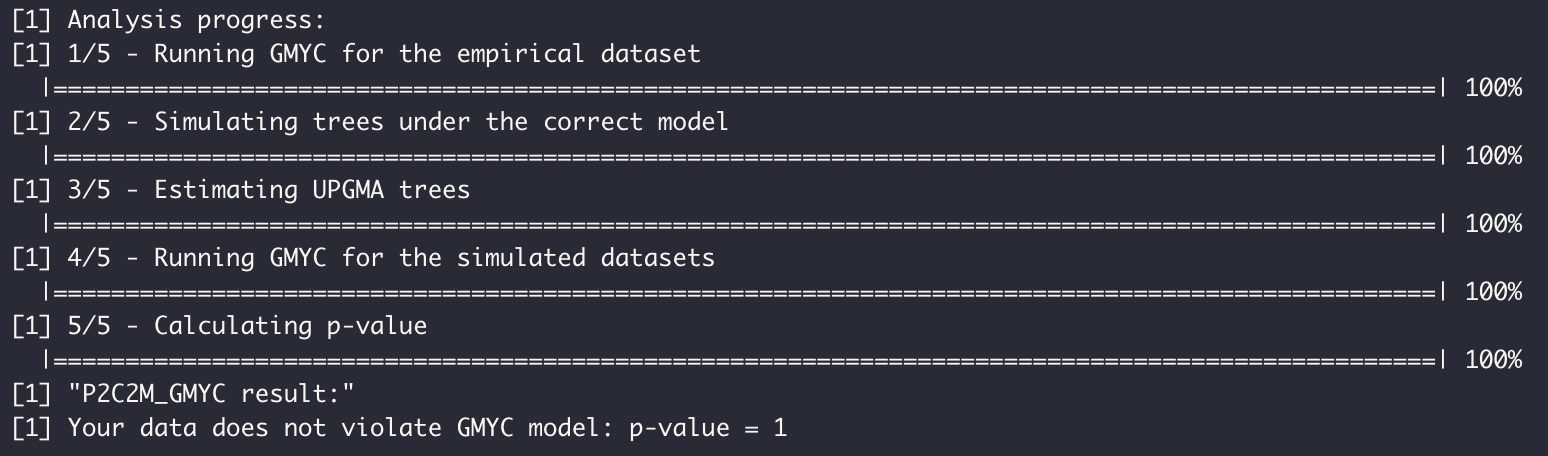
To run the parametric bootstrap version of the P2C2M.GMYC, users need to provide an ultrametric phylogenetic tree and a sequential fasta alignment.
We included Lygodactylus dataset in P2C2M.GMYC as an example. The Lygodactylus* dataset includes 66 sequences of the mitochondrial ND4 gene for 19 described species. This dataset was chosen for representing a typical dataset used in GMYC analysis, where each species can be represented from one to many sequences. So, we will investigate if the GMYC model is a good fit for the evolutionary history of Lygodactylus ssp. using the parametric bootstrap version.
library(P2C2M.GMYC)
P2C2M_GMYC.Pboot(tree.input=system.file("extdata", "Lygodactylus.tre", package="P2C2M.GMYC"),
tree.format="nexus",
seq=system.file("extdata", "Lygodactylus.fas", package="P2C2M.GMYC"))
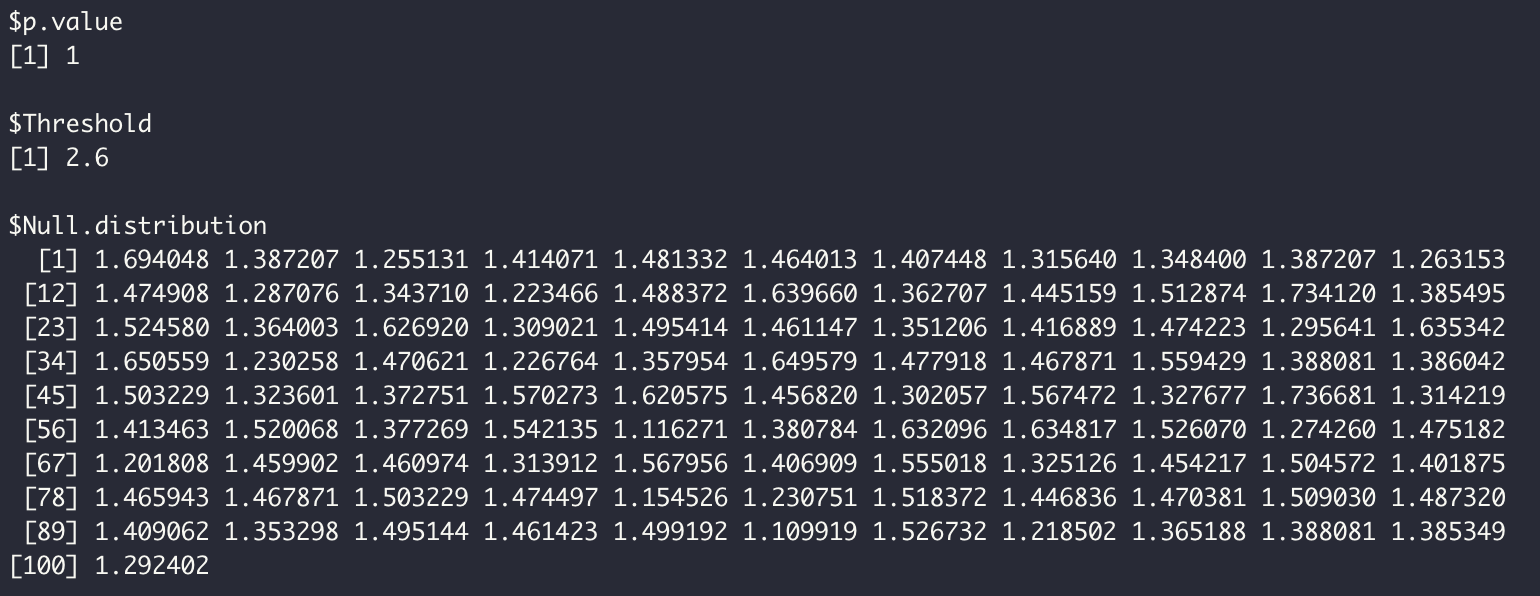
Results can be accessed in P2C2M_GMYC.results object.
P2C2M_GMYC.results
Reference:
Lanna, F.M., Werneck, F.P., Gehara, M., Fonseca, E.M., Colli, G.R., Sites-Jr., J.W., Rodrigues, M.T., and Garda, A.A. 2018. The evolutionary history of Lygodactylus lizards in the South American open diagonal. Molecular Phylogenetics and Evolution, 127: 638–645. https://doi.org/10.1016/j.ympev.2018.06.010.
Pons J., Barraclough, T.G., Gomez-Zurita, J., Cardoso, A., Duran, D.P., Hazel, S., Kamoun, S., Sumlin, W.D., Vogler, A.P. 2006. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Systematic Biology 55(3): 595-609.