Highly parallel DBSCAN (HPDBSCAN) is a shared- and distributed-memory parallel implementation of the Density-Based Spatial Clustering for Applications with Noise (DBSCAN) algorithm. It is written in C++ and may be used as shared library and command line tool.
HPDBSCAN requires the following dependencies. Please make sure, that these are installed, before attempting to compile the code.
- CMake 3.10+
- C++11 compliant compiler (e.g. g++ 4.9+)
- OpenMP 4.0+ (e.g. g++ 4.9+)
- HDF5 1.8+
- (optional) Message Passing Interface (MPI) 2.0+
- (optional) SWIG 3.0+ (Python bindings with and without MPI support)
- (optional) Python 3.5+ (Python bindings with and without MPI support, requires headers)
- (optional) mpi4py (Python bindings with MPI support)
HPDBSCAN follows the standard CMake project conventions. Create a build directory, change to it, generate the build script and compile it. A convenience short-hand can be found below.
mkdir build && cd build && cmake .. && makeThe provided CMake script checks, but does not install, all of the necessary dependencies listed above. If no MPI installation is present on the system, an OpenMP-only (i.e. thread-based) version is built.
HPDBSCAN's command line usage flags are shown below. You may obtain the same message by invoking hpdbscan -h:
Highly parallel DBSCAN clustering algorithm
Usage:
HPDBSCAN [OPTION...]
-h, --help this help message
-m, --minPoints arg density threshold (default: 2)
-e, --epsilon arg spatial search radius (default: 0.1)
-t, --threads arg utilized threads (default: 4)
-i, --input arg input file (default: data.h5)
-o, --output arg output file (default: data.h5)
--input-dataset arg input dataset name (default: DATA)
--output-dataset arg output dataset name (default: CLUSTERS)
The typical basic usage of HPDBSCAN is shown below. The above line is for single-node-only (e.g. your laptop/PC) execution. The line below shows a typical high-performance computing setup with multiple distributed nodes and processing cores per node. The data is passed to the application in form of an HDF5 file.
./hpdbscan -t <THREADS> <PATH_TO_HDF5_FILE>
mpirun -np <NODES> ./hpdbscan -t <THREADS> <PATH_TO_HDF5_FILE>A second file will be created containing a single vector with the labels for each data point at the same index. The labels may be unintuitive at first. The value zero indicates a noise point, labels unequal to zero indicate a point belonging to a cluster. Negative cluster values are core points of the respective cluster with the same absolute value.
For example, a point might have the cluster label -3, indicating it belongs to the cluster with ID 3 and it is a core point. A second might have the cluster label 3, indicating that it also belongs to the cluster with the ID 3, however, it is not a core point of said cluster. Nevertheless, all points with either the cluster labels -3 or 3 belong to the same cluster with the ID 3.
The CMake script will automatically build HPDBSCAN bindings for the Python programming language, if SWIG and Python (with headers) installation are found on the system. Additionally, if your Python installation also provides the mpi4py pip package, MPI support is enabled.
A small programming example is shown below:
import hpdbscan
import numpy as np
data = np.random.rand(100, 3)
clusterer = hpdbscan.HPDBSCAN(epsilon=0.3, min_points=4)
clusterer.cluster(data)
# alternatively
clusterer.cluster('file.h5', 'data')The data passed to the binding is expected to be a two-dimensional numpy array, where the first axis is the sample dimension and the second axis the feature dimension. The result is returned as a tuple with the same lenghts as the first data dimension.
Should you want to use the MPI flavor of the binding, please ensure that each MPI rank only receives a disjoint subset of the entire dataset (e.g. equally-sized chunks). After the clustering process each rank will have the labels corresponding to the initially passed data items.
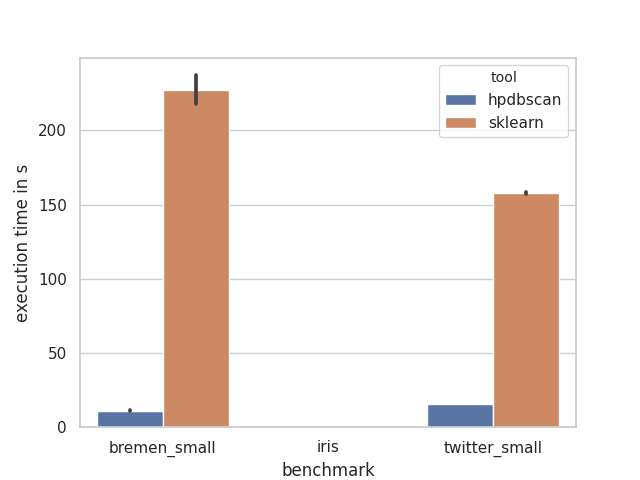
Based on empirical benchmarks HPDBSCAN outperforms other DBSCAN implementation by a signficant margin in terms of execution time. One benchmarking review has been conducted by Helmut Neukirchen [1] for example. Beyond that, the repository contains a small benchmarking suite. If you want to redo them, please ensure that you have a Python 3.6+ interpreter and the pip packages numpy, pandas, sklearn, h5py, seaborn installed and execute the scripts in this order: 1. download_datasets.py, 2. benchmark.py and 3. plot.py.
Below you will find a figure with an execution of the benchmark suite. For each of the three presented datasets, 10 execution time measurement runs have been performed. The plot depicts the average of the execution time and the black bars the standard deviation, i.e. the execution time fluctuation. All runs have been performed on an single server with an Intel Xeon Gold 6126. The thread count was set to 24, the number of hardware cores, for both tools. The numpy used in sklearn is linked against the Intel MKL 2019.1.
The benchmark run for iris is not a mistake, it just executes subsecond for both tools and is therefore barely visible.
[1] Neukirchen, Helmut. Survey and performance evaluation of DBSCAN spatial clustering implementations for big data and high-performance computing paradigms. Technical report VHI-01-2016, Engineering Research Institute, University of Iceland, 2016.
If you wish to cite HPDBSCAN in your academic work, please use the following reference:
Plain reference
Götz, M., Bodenstein, C., Riedel M.,
HPDBSCAN: highly parallel DBSCAN,
Proceedings of the Workshop on Machine Learning in High-Performance Computing Environments, ACM, 2015.
BibTex
@inproceedings{gotz2015hpdbscan,
title={HPDBSCAN: highly parallel DBSCAN},
author={G{\"o}tz, Markus and Bodenstein, Christian and Riedel, Morris},
booktitle={Proceedings of the Workshop on Machine Learning in High-Performance Computing Environments},
pages={2},
year={2015},
organization={ACM}
}If you want to let us know about feature requests, bugs or issues you are kindly referred to the issue tracker.
For any other discussion, please write an e-mail.