Neural cellular automata (NCA) represent a remarkable fusion of neural networks and cellular automata, enabling the simulation of complex systems and showcasing fascinating emergent behavior. In this project, we investigated their capabilities in the field of medical imaging. Specifically, we train NCA models on the MedMNIST dataset. Our implementation is based on the code in the paper Growing Neural Cellular Automata. We provide code for training NCA models using different filter and loss functions, visualizing the dynamic creation of an image based on a pre-trained model, and investigating the influence of various hyperparameters.
$ git clone https://github.com/Marlon154/DGM-Project-NCA.git
$ cd DGM-Project_NCA
docker build -f Dockerfile -t dgm .
To train an NCA model, run the docker image you just built:
docker run -it dgm
The config.yaml allows you to set various hyperparameters, including which filters and losses to use. The following are available:
Filters: [sobel, laplacian, gaussian, sobel_identity] + _identity
Losses: mse, manhattan, hinge, ssim, combined_ssim_l1
Our investigation showed that all of our filters only work well in combination with the identity filter. Therefore, when, for example, using the Sobel filter, append the string "_identity" in the hyperparameter settings (e.g., "sobel_identity"). The resulting model will be saved in the models/ folder.
To visualize the creation of an image, run the create_all_gifs.py file. This will create a GIF for each model in your model/ folder.
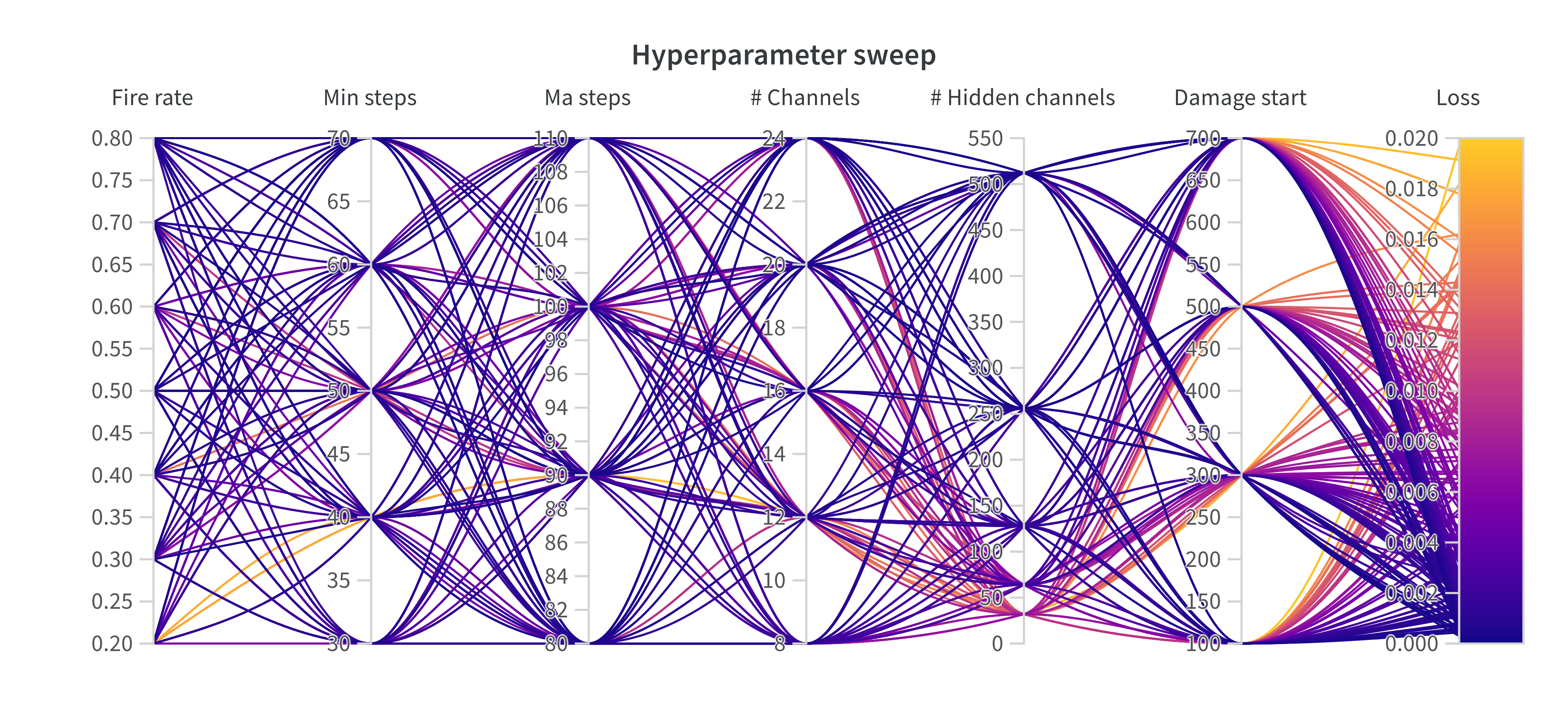
Additionally, we provide code for conducting a hyperparameter sweep, which allows an investigation into which hyperparameters are most important and what values work best. We use the sweep protocol provided by wandb. To conduct a hyperparameter sweep, run the hyperparameter_tuning.py file. To allow additional supervision on the parameters included in the sweep we provided a sweep_config.yaml file. Following, you can see an example visualization of a sweep:
You can use the script download_images.py in the src folder to download the images.
The script requires a file with the images' URLs to download.
The ca{{ should have the following format:
python src/download_images.py bloodmnist --size 28 --split test --index 0
Possible datasets: 'pathmnist', 'chestmnist', 'dermamnist', 'octmnist', 'pneumoniamnist', 'retinamnist', 'breastmnist', 'bloodmnist', 'tissuemnist', 'organamnist', 'organcmnist', 'organsmnist'
For the final presentation, we created a scientific poster summarizing our work, which can be seen here: Poster