Multi-View GCN (MVGCN) for Brain Networks
Overview
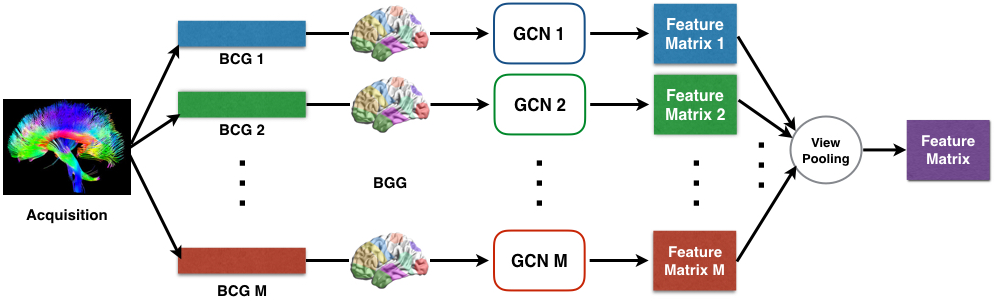
This repository contains TensorFlow code for implementing Multi-View Graph Convolutional Network for brain networks (DTI data). To do this, we train models using graph convolutional networks (GCNs) in multiple views to learn view-based feature representations. Then, view pooling is conducted for the purpose of multi-view feature fusion. The code is documented and should be easy to modify for your own applications.
Model
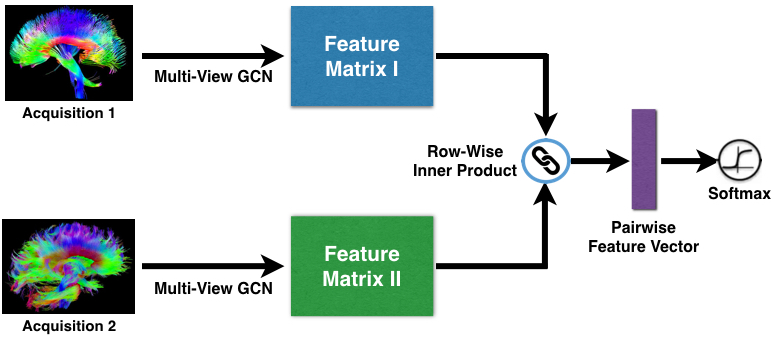
The objective function is established for a binary classification problem, which is matching vs. non-matching classes. Brain networks in the same group (Parkinson's Disease or Healthy Control) are labeled as matching pairs while brain networks from different groups are labeled as non-matching pairs. Hence, pairwise training samples are feed into the neural network. The figure depicts the pairwise learning architecture.
- The components utilized in the MVGCN are:
- GCNs: incorporating coordinates of ROI (Region of Interest) with brain networks (DTI) together to learn interpretable representations.
- View pooling: combining view-based representations of DTI data that are obtained from various tractography algorithms.
- Pairwise matching: computing similarities by euclidean distance for sample pairs in terms of the pooled features.
- Softmax: classifying the output of pairwise matching layer into the matching vs. non-matching classes.
This repository contains the slides we presented in AMIA 2018.
Requirements
This package has the following requirements:
- An NVIDIA GPU.
Python 3.x- TensorFlow 1.4
Usage
How to Run
To run MVGCN on your data, you need to: change the function of loading data in utils.py; define the names of multiple views in mvgcn.sh; set hyperparameters for MVGCN in mvgcn.sh; run the shell script mvgcn.sh
bash mvgcn.shData Format
The input file "dti.coo.pkl" is the XYZ-coordinate for given samples with the 3d dimension: n_samples x n_nodes x n_XYZ, where n_node equals to the number of ROI in our case. n_XYZ equals to 3 which indicates the X axis, Y axis, and Z axis.
The input file "dti.pair.pkl" stores the index of samples and their corresponding labels by a tuple(pair, label), where pair is a list of pairwise indices. And label is a list of integer values. Here is an example with several sample pairs and labels:
| Index 1 | Index 2 | Label |
|---|---|---|
| 0 | 8 | 1 |
| 0 | 9 | 1 |
| 0 | 10 | 1 |
| 0 | 26 | 0 |
| 1 | 5 | 0 |
| 1 | 6 | 0 |
| ... | ... | ... |
In detail, we use label 1 to indicate the sample pair belongs to the same class (matching) and label 0 to indicate it belongs to the different classes (non-matching). The index begins from 0.
Additional Material
There is implementations used in:
Michaël Defferrard, Xavier Bresson, Pierre Vandergheynst, Convolutional Neural Networks on Graphs with Fast Localized Spectral Filtering, Neural Information Processing Systems (NIPS), 2016.
Sofia Ira Ktena, Sarah Parisot, Enzo Ferrante, Martin Rajchl, Matthew Lee, Ben Glocker, Daniel Rueckert, Distance Metric Learning using Graph Convolutional Networks: Application to Functional Brain Networks, Medical Image Computing and Computer-Assisted Interventions (MICCAI), 2017.
References
If you happen to use our work, please consider citing our paper:
@article{zhang2018multi,
title={Multi-View Graph Convolutional Network and Its Applications on Neuroimage Analysis for Parkinson's Disease},
author={Zhang, Xi and He, Lifang and Chen, Kun and Luo, Yuan and Zhou, Jiayu and Wang, Fei},
journal={arXiv preprint arXiv:1805.08801},
year={2018}
}
This paper can be accessed on : [Multi-View GCN] (https://arxiv.org/pdf/1805.08801.pdf)
Acknowledgements
We owe many thanks to Dr. Liang Zhan for helping us on processing the neuroimages. Please see the paper for funding details and additional (non-code related) acknowledgements.